Ribosome profiling reveals sequence-independent post-initiation pausing as a signature of translation
- PMID: 24903108
- PMCID: PMC4085768
- DOI: 10.1038/cr.2014.74
Ribosome profiling reveals sequence-independent post-initiation pausing as a signature of translation
Abstract
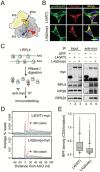
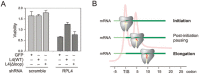
The journey of a newly synthesized polypeptide starts in the peptidyltransferase center of the ribosome, from where it traverses the exit tunnel. The interior of the ribosome exit tunnel is neither straight nor smooth. How the ribosome dynamics in vivo is influenced by the exit tunnel is poorly understood. Genome-wide ribosome profiling in mammalian cells reveals elevated ribosome density at the start codon and surprisingly the downstream 5th codon position as well. We found that the highly focused ribosomal pausing shortly after initiation is attributed to the geometry of the exit tunnel, as deletion of the loop region from ribosome protein L4 diminishes translational pausing at the 5th codon position. Unexpectedly, the ribosome variant undergoes translational abandonment shortly after initiation, suggesting that there exists an obligatory step between initiation and elongation commitment. We propose that the post-initiation pausing of ribosomes represents an inherent signature of the translation machinery to ensure productive translation.
Figures


References
-
- Buchan JR, Stansfield I. Halting a cellular production line: responses to ribosomal pausing during translation. Biol Cell. 2007;99:475–487. - PubMed
-
- Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–597. - PubMed
-
- Farabaugh PJ. Translational frameshifting: implications for the mechanism of translational frame maintenance. Prog Nucleic Acid Res Mol Biol. 2000;64:131–170. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

