Bears in a forest of gene trees: phylogenetic inference is complicated by incomplete lineage sorting and gene flow
- PMID: 24903145
- PMCID: PMC4104321
- DOI: 10.1093/molbev/msu186
Bears in a forest of gene trees: phylogenetic inference is complicated by incomplete lineage sorting and gene flow
Abstract
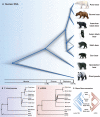
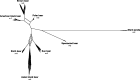
Ursine bears are a mammalian subfamily that comprises six morphologically and ecologically distinct extant species. Previous phylogenetic analyses of concatenated nuclear genes could not resolve all relationships among bears, and appeared to conflict with the mitochondrial phylogeny. Evolutionary processes such as incomplete lineage sorting and introgression can cause gene tree discordance and complicate phylogenetic inferences, but are not accounted for in phylogenetic analyses of concatenated data. We generated a high-resolution data set of autosomal introns from several individuals per species and of Y-chromosomal markers. Incorporating intraspecific variability in coalescence-based phylogenetic and gene flow estimation approaches, we traced the genealogical history of individual alleles. Considerable heterogeneity among nuclear loci and discordance between nuclear and mitochondrial phylogenies were found. A species tree with divergence time estimates indicated that ursine bears diversified within less than 2 My. Consistent with a complex branching order within a clade of Asian bear species, we identified unidirectional gene flow from Asian black into sloth bears. Moreover, gene flow detected from brown into American black bears can explain the conflicting placement of the American black bear in mitochondrial and nuclear phylogenies. These results highlight that both incomplete lineage sorting and introgression are prominent evolutionary forces even on time scales up to several million years. Complex evolutionary patterns are not adequately captured by strictly bifurcating models, and can only be fully understood when analyzing multiple independently inherited loci in a coalescence framework. Phylogenetic incongruence among gene trees hence needs to be recognized as a biologically meaningful signal.
Keywords: Ursidae; coalescence; introgressive hybridization; multi-locus analyses; phylogenetic network; species tree.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

Similar articles
-
Rapid radiation events in the family Ursidae indicated by likelihood phylogenetic estimation from multiple fragments of mtDNA.Mol Phylogenet Evol. 1999 Oct;13(1):82-92. doi: 10.1006/mpev.1999.0637. Mol Phylogenet Evol. 1999. PMID: 10508542
-
Brown and polar bear Y chromosomes reveal extensive male-biased gene flow within brother lineages.Mol Biol Evol. 2014 Jun;31(6):1353-63. doi: 10.1093/molbev/msu109. Epub 2014 Mar 25. Mol Biol Evol. 2014. PMID: 24667925
-
Phylogenetic Conflict in Bears Identified by Automated Discovery of Transposable Element Insertions in Low-Coverage Genomes.Genome Biol Evol. 2017 Oct 1;9(10):2862-2878. doi: 10.1093/gbe/evx170. Genome Biol Evol. 2017. PMID: 28985298 Free PMC article.
-
Understanding phylogenetic incongruence: lessons from phyllostomid bats.Biol Rev Camb Philos Soc. 2012 Nov;87(4):991-1024. doi: 10.1111/j.1469-185X.2012.00240.x. Epub 2012 Aug 14. Biol Rev Camb Philos Soc. 2012. PMID: 22891620 Free PMC article. Review.
-
Reticulate evolution: Detection and utility in the phylogenomics era.Mol Phylogenet Evol. 2024 Dec;201:108197. doi: 10.1016/j.ympev.2024.108197. Epub 2024 Sep 11. Mol Phylogenet Evol. 2024. PMID: 39270765 Review.
Cited by
-
Maintenance of Species Boundaries Despite Ongoing Gene Flow in Ragworts.Genome Biol Evol. 2016 Apr 13;8(4):1038-47. doi: 10.1093/gbe/evw053. Genome Biol Evol. 2016. PMID: 26979797 Free PMC article.
-
Phylogenomic Signatures of Ancient Introgression in a Rogue Lineage of Darters (Teleostei: Percidae).Syst Biol. 2019 Mar 1;68(2):329-346. doi: 10.1093/sysbio/syy074. Syst Biol. 2019. PMID: 30395332 Free PMC article.
-
Contrasting responses to Pleistocene climate changes: a case study of two sister species Allium cyathophorum and A. spicata (Amaryllidaceae) distributed in the eastern and western Qinghai-Tibet Plateau.Ecol Evol. 2015 Apr;5(7):1513-24. doi: 10.1002/ece3.1449. Epub 2015 Mar 10. Ecol Evol. 2015. PMID: 25897390 Free PMC article.
-
Additive Traits Lead to Feeding Advantage and Reproductive Isolation, Promoting Homoploid Hybrid Speciation.Mol Biol Evol. 2019 Aug 1;36(8):1671-1685. doi: 10.1093/molbev/msz090. Mol Biol Evol. 2019. PMID: 31028398 Free PMC article.
-
Multilocus Intron Trees Reveal Extensive Male-Biased Homogenization of Ancient Populations of Chamois (Rupicapra spp.) across Europe during Late Pleistocene.PLoS One. 2017 Feb 1;12(2):e0170392. doi: 10.1371/journal.pone.0170392. eCollection 2017. PLoS One. 2017. PMID: 28146581 Free PMC article.
References
-
- Bapteste E, van Iersel L, Janke A, Kelchner S, Kelk S, McInerney JO, Morrison DA, Nakhleh L, Steel M, Stougie L, et al. Networks: expanding evolutionary thinking. Trends Genet. 2013;29:439–441. - PubMed
-
- Bidon T, Janke A, Fain SR, Eiken HG, Hagen SB, Saarma U, Hallström BM, Lecomte N, Hailer F. Brown and polar bear Y chromosomes reveal extensive male-biased gene flow within brother lineages. Mol Biol Evol. 2014;31:1353–1363. - PubMed
-
- Bonner JT. Size and cycles: an essay on the structure of biology. Princeton (NY): Princeton University Press; 1965.
-
- Bouckaert RR. DensiTree: making sense of sets of phylogenetic trees. Bioinformatics. 2010;26:1372–1373. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

