A novel atypical hemolytic uremic syndrome-associated hybrid CFHR1/CFH gene encoding a fusion protein that antagonizes factor H-dependent complement regulation
- PMID: 24904082
- PMCID: PMC4279739
- DOI: 10.1681/ASN.2013121339
A novel atypical hemolytic uremic syndrome-associated hybrid CFHR1/CFH gene encoding a fusion protein that antagonizes factor H-dependent complement regulation
Abstract
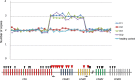
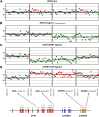
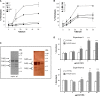
Genomic aberrations affecting the genes encoding factor H (FH) and the five FH-related proteins (FHRs) have been described in patients with atypical hemolytic uremic syndrome (aHUS), a rare condition characterized by microangiopathic hemolytic anemia, thrombocytopenia, and ARF. These genomic rearrangements occur through nonallelic homologous recombinations caused by the presence of repeated homologous sequences in CFH and CFHR1-R5 genes. In this study, we found heterozygous genomic rearrangements among CFH and CFHR genes in 4.5% of patients with aHUS. CFH/CFHR rearrangements were associated with poor clinical prognosis and high risk of post-transplant recurrence. Five patients carried known CFH/CFHR1 genes, but we found a duplication leading to a novel CFHR1/CFH hybrid gene in a family with two affected subjects. The resulting fusion protein contains the first four short consensus repeats of FHR1 and the terminal short consensus repeat 20 of FH. In an FH-dependent hemolysis assay, we showed that the hybrid protein causes sheep erythrocyte lysis. Functional analysis of the FHR1 fraction purified from serum of heterozygous carriers of the CFHR1/CFH hybrid gene indicated that the FHR1/FH hybrid protein acts as a competitive antagonist of FH. Furthermore, sera from carriers of the hybrid CFHR1/CFH gene induced more C5b-9 deposition on endothelial cells than control serum. These results suggest that this novel genomic hybrid mediates disease pathogenesis through dysregulation of complement at the endothelial cell surface. We recommend that genetic screening of aHUS includes analysis of CFH and CFHR rearrangements, particularly before a kidney transplant.
Keywords: complement; genetic renal disease; hemolytic uremic syndrome; kidney disease; transplantation.
Copyright © 2015 by the American Society of Nephrology.
Figures




References
-
- Noris M, Remuzzi G: Atypical hemolytic-uremic syndrome. N Engl J Med 361: 1676–1687, 2009 - PubMed
-
- Kavanagh D, Goodship TH: Atypical hemolytic uremic syndrome. Curr Opin Hematol 17: 432–438, 2010 - PubMed
-
- Bresin E, Rurali E, Caprioli J, Sanchez-Corral P, Fremeaux-Bacchi V, Rodriguez de Cordoba S, Pinto S, Goodship TH, Alberti M, Ribes D, Valoti E, Remuzzi G, Noris M, European Working Party on Complement Genetics in Renal Diseases : Combined complement gene mutations in atypical hemolytic uremic syndrome influence clinical phenotype. J Am Soc Nephrol 24: 475–486, 2013 - PMC - PubMed
-
- Dragon-Durey MA, Loirat C, Cloarec S, Macher MA, Blouin J, Nivet H, Weiss L, Fridman WH, Frémeaux-Bacchi V: Anti-Factor H autoantibodies associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol 16: 555–563, 2005 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

