Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions
- PMID: 24904623
- PMCID: PMC4036129
- DOI: 10.3389/fpls.2014.00228
Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions
Abstract
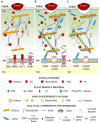
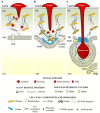
The cell wall is a dynamic structure that often determines the outcome of the interactions between plants and pathogens. It is a barrier that pathogens need to breach to colonize the plant tissue. While fungal necrotrophs extensively destroy the integrity of the cell wall through the combined action of degrading enzymes, biotrophic fungi require a more localized and controlled degradation of the cell wall in order to keep the host cells alive and utilize their feeding structures. Also bacteria and nematodes need to degrade the plant cell wall at a certain stage of their infection process, to obtain nutrients for their growth. Plants have developed a system for sensing pathogens and monitoring the cell wall integrity, upon which they activate defense responses that lead to a dynamic cell wall remodeling required to prevent the disease. Pathogens, on the other hand, may exploit the host cell wall metabolism to support the infection. We review here the strategies utilized by both plants and pathogens to prevail in the cell wall battleground.
Keywords: cell wall; cell wall integrity; host cell wall metabolism reprogramming; plant defense; susceptibility factors.
Figures
References
-
- Aguero C. B., Uratsu S. L., Greve C., Powell A. L. T., Labavitch J. M., Meredith C. P., et al. (2005). Evaluation of tolerance to pierce’s disease and botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol. Plant Pathol. 6 43–51 10.1111/j.1364-3703.2004.00262.x - DOI - PubMed
-
- Aist J. R. (1976). Papillae and related wound plugs of plant cells. Annu. Rev. Phytopathol. 14 145–163 10.1146/annurev.py.14.090176.001045 - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

