Metabolomic tools for secondary metabolite discovery from marine microbial symbionts
- PMID: 24905482
- PMCID: PMC4071584
- DOI: 10.3390/md12063416
Metabolomic tools for secondary metabolite discovery from marine microbial symbionts
Abstract
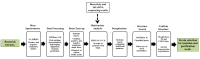
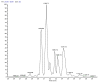
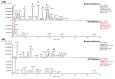
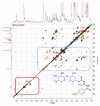
Marine invertebrate-associated symbiotic bacteria produce a plethora of novel secondary metabolites which may be structurally unique with interesting pharmacological properties. Selection of strains usually relies on literature searching, genetic screening and bioactivity results, often without considering the chemical novelty and abundance of secondary metabolites being produced by the microorganism until the time-consuming bioassay-guided isolation stages. To fast track the selection process, metabolomic tools were used to aid strain selection by investigating differences in the chemical profiles of 77 bacterial extracts isolated from cold water marine invertebrates from Orkney, Scotland using liquid chromatography-high resolution mass spectrometry (LC-HRMS) and nuclear magnetic resonance (NMR) spectroscopy. Following mass spectrometric analysis and dereplication using an Excel macro developed in-house, principal component analysis (PCA) was employed to differentiate the bacterial strains based on their chemical profiles. NMR 1H and correlation spectroscopy (COSY) were also employed to obtain a chemical fingerprint of each bacterial strain and to confirm the presence of functional groups and spin systems. These results were then combined with taxonomic identification and bioassay screening data to identify three bacterial strains, namely Bacillus sp. 4117, Rhodococcus sp. ZS402 and Vibrio splendidus strain LGP32, to prioritize for scale-up based on their chemically interesting secondary metabolomes, established through dereplication and interesting bioactivities, determined from bioassay screening.
Figures







Similar articles
-
Metabolomics and dereplication strategies in natural products.Methods Mol Biol. 2013;1055:227-44. doi: 10.1007/978-1-62703-577-4_17. Methods Mol Biol. 2013. PMID: 23963915
-
Chemical dereplication of marine actinomycetes by liquid chromatography-high resolution mass spectrometry profiling and statistical analysis.Anal Chim Acta. 2013 Dec 17;805:70-9. doi: 10.1016/j.aca.2013.10.029. Epub 2013 Oct 21. Anal Chim Acta. 2013. PMID: 24296145
-
Chemical screening method for the rapid identification of microbial sources of marine invertebrate-associated metabolites.Mar Drugs. 2011 Mar 21;9(3):369-81. doi: 10.3390/md9030369. Mar Drugs. 2011. PMID: 21556166 Free PMC article.
-
Metabolomic Strategies Involving Mass Spectrometry Combined with Liquid and Gas Chromatography.Adv Exp Med Biol. 2017;965:77-98. doi: 10.1007/978-3-319-47656-8_4. Adv Exp Med Biol. 2017. PMID: 28132177 Review.
-
Beyond the paradigm: Combining mass spectrometry and nuclear magnetic resonance for metabolomics.Prog Nucl Magn Reson Spectrosc. 2017 May;100:1-16. doi: 10.1016/j.pnmrs.2017.01.001. Epub 2017 Jan 11. Prog Nucl Magn Reson Spectrosc. 2017. PMID: 28552170 Free PMC article. Review.
Cited by
-
Multistage Detection of Tetrodotoxin Traces in Diodon hystrix Collected in El Salvador.Toxins (Basel). 2023 Jun 25;15(7):409. doi: 10.3390/toxins15070409. Toxins (Basel). 2023. PMID: 37505678 Free PMC article.
-
Using Molecular Networking for Microbial Secondary Metabolite Bioprospecting.Metabolites. 2016 Jan 8;6(1):2. doi: 10.3390/metabo6010002. Metabolites. 2016. PMID: 26761036 Free PMC article.
-
A metabolomic approach to target antimalarial metabolites in the Artemisia annua fungal endophytes.Sci Rep. 2021 Feb 2;11(1):2770. doi: 10.1038/s41598-021-82201-8. Sci Rep. 2021. PMID: 33531542 Free PMC article.
-
Defining the biomarkers in anti-MRSA fractions of soil Streptomycetes by multivariate analysis.Antonie Van Leeuwenhoek. 2025 May 10;118(6):77. doi: 10.1007/s10482-025-02087-8. Antonie Van Leeuwenhoek. 2025. PMID: 40347296
-
hcapca: Automated Hierarchical Clustering and Principal Component Analysis of Large Metabolomic Datasets in R.Metabolites. 2020 Jul 21;10(7):297. doi: 10.3390/metabo10070297. Metabolites. 2020. PMID: 32708222 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

