Cell cycle-regulated transcription: effectively using a genomics toolbox
- PMID: 24906306
- PMCID: PMC7123442
- DOI: 10.1007/978-1-4939-0888-2_1
Cell cycle-regulated transcription: effectively using a genomics toolbox
Abstract
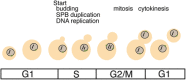
The cell cycle comprises a series of temporally ordered events that occur sequentially, including DNA replication, centrosome duplication, mitosis, and cytokinesis. What are the regulatory mechanisms that ensure proper timing and coordination of events during the cell cycle? Biochemical and genetic screens have identified a number of cell-cycle regulators, and it was recognized early on that many of the genes encoding cell-cycle regulators, including cyclins, were transcribed only in distinct phases of the cell cycle. Thus, "just in time" expression is likely an important part of the mechanism that maintains the proper temporal order of cell cycle events. New high-throughput technologies for measuring transcript levels have revealed that a large percentage of the Saccharomyces cerevisiae transcriptome (~20 %) is cell cycle regulated. Similarly, a substantial fraction of the mammalian transcriptome is cell cycle-regulated. Over the past 25 years, many studies have been undertaken to determine how gene expression is regulated during the cell cycle. In this review, we discuss contemporary models for the control of cell cycle-regulated transcription, and how this transcription program is coordinated with other cell cycle events in S. cerevisiae. In addition, we address the genomic approaches and analytical methods that enabled contemporary models of cell cycle transcription. Finally, we address current and future technologies that will aid in further understanding the role of periodic transcription during cell cycle progression.
Figures

References
-
- Morgan DO. The cell cycle: principles of control. Primers in biology. London: New Science Press in association with Oxford University Press; 2007.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

