Profiling the orphan enzymes
- PMID: 24906382
- PMCID: PMC4084501
- DOI: 10.1186/1745-6150-9-10
Profiling the orphan enzymes
Abstract
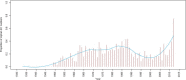
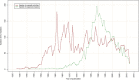
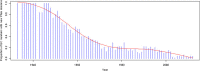
The emergence of Next Generation Sequencing generates an incredible amount of sequence and great potential for new enzyme discovery. Despite this huge amount of data and the profusion of bioinformatic methods for function prediction, a large part of known enzyme activities is still lacking an associated protein sequence. These particular activities are called "orphan enzymes". The present review proposes an update of previous surveys on orphan enzymes by mining the current content of public databases. While the percentage of orphan enzyme activities has decreased from 38% to 22% in ten years, there are still more than 1,000 orphans among the 5,000 entries of the Enzyme Commission (EC) classification. Taking into account all the reactions present in metabolic databases, this proportion dramatically increases to reach nearly 50% of orphans and many of them are not associated to a known pathway. We extended our survey to "local orphan enzymes" that are activities which have no representative sequence in a given clade, but have at least one in organisms belonging to other clades. We observe an important bias in Archaea and find that in general more than 30% of the EC activities have incomplete sequence information in at least one superkingdom. To estimate if candidate proteins for local orphans could be retrieved by homology search, we applied a simple strategy based on the PRIAM software and noticed that candidates may be proposed for an important fraction of local orphan enzymes. Finally, by studying relation between protein domains and catalyzed activities, it appears that newly discovered enzymes are mostly associated with already known enzyme domains. Thus, the exploration of the promiscuity and the multifunctional aspect of known enzyme families may solve part of the orphan enzyme issue. We conclude this review with a presentation of recent initiatives in finding proteins for orphan enzymes and in extending the enzyme world by the discovery of new activities.
Figures




References
-
- Payen A, Perzoz J. Mémoire sur la diastase, les principaux produits de ses rèactions, et leurs applications aux arts industriels. Annales de la chimie et de la physique. 1833;53:73–92.
-
- Tipton K, Boyce S. History of the enzyme nomenclature system. Bioinformatics. 2000;16:34–40. - PubMed
-
- Enzyme nomenclature. http://www.chem.qmul.ac.uk/iubmb/enzyme/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

