Population-specific common SNPs reflect demographic histories and highlight regions of genomic plasticity with functional relevance
- PMID: 24906912
- PMCID: PMC4092225
- DOI: 10.1186/1471-2164-15-437
Population-specific common SNPs reflect demographic histories and highlight regions of genomic plasticity with functional relevance
Abstract
Background: Population differentiation is the result of demographic and evolutionary forces. Whole genome datasets from the 1000 Genomes Project (October 2012) provide an unbiased view of genetic variation across populations from Europe, Asia, Africa and the Americas. Common population-specific SNPs (MAF > 0.05) reflect a deep history and may have important consequences for health and wellbeing. Their interpretation is contextualised by currently available genome data.
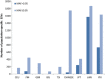
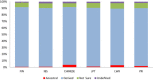
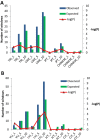
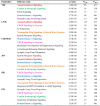
Results: The identification of common population-specific (CPS) variants (SNPs and SSV) is influenced by admixture and the sample size under investigation. Nine of the populations in the 1000 Genomes Project (2 African, 2 Asian (including a merged Chinese group) and 5 European) revealed that the African populations (LWK and YRI), followed by the Japanese (JPT) have the highest number of CPS SNPs, in concordance with their histories and given the populations studied. Using two methods, sliding 50-SNP and 5-kb windows, the CPS SNPs showed distinct clustering across large genome segments and little overlap of clusters between populations. iHS enrichment score and the population branch statistic (PBS) analyses suggest that selective sweeps are unlikely to account for the clustering and population specificity. Of interest is the association of clusters close to recombination hotspots. Functional analysis of genes associated with the CPS SNPs revealed over-representation of genes in pathways associated with neuronal development, including axonal guidance signalling and CREB signalling in neurones.
Conclusions: Common population-specific SNPs are non-randomly distributed throughout the genome and are significantly associated with recombination hotspots. Since the variant alleles of most CPS SNPs are the derived allele, they likely arose in the specific population after a split from a common ancestor. Their proximity to genes involved in specific pathways, including neuronal development, suggests evolutionary plasticity of selected genomic regions. Contrary to expectation, selective sweeps did not play a large role in the persistence of population-specific variation. This suggests a stochastic process towards population-specific variation which reflects demographic histories and may have some interesting implications for health and susceptibility to disease.
Figures




Similar articles
-
Genome-wide association study SNPs in the human genome diversity project populations: does selection affect unlinked SNPs with shared trait associations?PLoS Genet. 2011 Jan 6;7(1):e1001266. doi: 10.1371/journal.pgen.1001266. PLoS Genet. 2011. PMID: 21253569 Free PMC article.
-
Similarity in recombination rate and linkage disequilibrium at CYP2C and CYP2D cytochrome P450 gene regions among Europeans indicates signs of selection and no advantage of using tagSNPs in population isolates.Pharmacogenet Genomics. 2012 Dec;22(12):846-57. doi: 10.1097/FPC.0b013e32835a3a6d. Pharmacogenet Genomics. 2012. PMID: 23089684
-
A whole genome long-range haplotype (WGLRH) test for detecting imprints of positive selection in human populations.Bioinformatics. 2006 Sep 1;22(17):2122-8. doi: 10.1093/bioinformatics/btl365. Epub 2006 Jul 15. Bioinformatics. 2006. PMID: 16845142
-
SNP ascertainment bias in population genetic analyses: why it is important, and how to correct it.Bioessays. 2013 Sep;35(9):780-6. doi: 10.1002/bies.201300014. Epub 2013 Jul 9. Bioessays. 2013. PMID: 23836388 Free PMC article. Review.
-
Advancements and prospects in reconstructing the genetic genealogies of ancient and modern human populations using ancestral recombination graphs.Yi Chuan. 2024 Oct;46(10):849-859. doi: 10.16288/j.yczz.24-150. Yi Chuan. 2024. PMID: 39443313 Review.
Cited by
-
Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program.Nature. 2021 Feb;590(7845):290-299. doi: 10.1038/s41586-021-03205-y. Epub 2021 Feb 10. Nature. 2021. PMID: 33568819 Free PMC article.
-
A novel locus in CSMD1 gene is associated with increased susceptibility to severe malaria in Malian children.Front Genet. 2024 May 24;15:1390786. doi: 10.3389/fgene.2024.1390786. eCollection 2024. Front Genet. 2024. PMID: 38854427 Free PMC article.
-
Genetics of autoimmune diseases: insights from population genetics.J Hum Genet. 2015 Nov;60(11):657-64. doi: 10.1038/jhg.2015.94. Epub 2015 Jul 30. J Hum Genet. 2015. PMID: 26223182 Free PMC article. Review.
-
High light intensity plays a major role in emergence of population level variation in Arabidopsis thaliana along an altitudinal gradient.Sci Rep. 2016 May 23;6:26160. doi: 10.1038/srep26160. Sci Rep. 2016. PMID: 27211014 Free PMC article.
-
Clinical and genetic insights of Parkinson's Disease in a Mexican cohort: highlighting Latino's diversity.medRxiv [Preprint]. 2025 May 3:2023.08.28.23294700. doi: 10.1101/2023.08.28.23294700. medRxiv. 2025. PMID: 37693616 Free PMC article. Preprint.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

