Chromatin organization and remodeling of interstitial telomeric sites during meiosis in the Mongolian gerbil (Meriones unguiculatus)
- PMID: 24907260
- PMCID: PMC4125389
- DOI: 10.1534/genetics.114.166421
Chromatin organization and remodeling of interstitial telomeric sites during meiosis in the Mongolian gerbil (Meriones unguiculatus)
Abstract
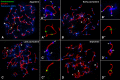
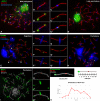
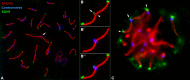
Telomeric DNA repeats are key features of chromosomes that allow the maintenance of integrity and stability in the telomeres. However, interstitial telomere sites (ITSs) can also be found along the chromosomes, especially near the centromere, where they may appear following chromosomal rearrangements like Robertsonian translocations. There is no defined role for ITSs, but they are linked to DNA damage-prone sites. We were interested in studying the structural organization of ITSs during meiosis, a kind of cell division in which programmed DNA damage events and noticeable chromatin reorganizations occur. Here we describe the presence of highly amplified ITSs in the pericentromeric region of Mongolian gerbil (Meriones unguiculatus) chromosomes. During meiosis, ITSs show a different chromatin conformation than DNA repeats at telomeres, appearing more extended and accumulating heterochromatin markers. Interestingly, ITSs also recruit the telomeric proteins RAP1 and TRF1, but in a stage-dependent manner, appearing mainly at late prophase I stages. We did not find a specific accumulation of DNA repair factors to the ITSs, such as γH2AX or RAD51 at these stages, but we could detect the presence of MLH1, a marker for reciprocal recombination. However, contrary to previous reports, we did not find a specific accumulation of crossovers at ITSs. Intriguingly, some centromeric regions of metacentric chromosomes may bind the nuclear envelope through the association to SUN1 protein, a feature usually performed by telomeres. Therefore, ITSs present a particular and dynamic chromatin configuration in meiosis, which could be involved in maintaining their genetic stability, but they additionally retain some features of distal telomeres, provided by their capability to associate to telomere-binding proteins.
Keywords: Mongolian gerbil; RAP1; TRF1; chromatin; interstitial telomeres; meiosis.
Copyright © 2014 by the Genetics Society of America.
Figures





References
-
- Abuín M., Martínez P., Sánchez L., 1996. Localization of the repetitive telomeric sequence (TTAGGG)n in four salmonid species. Genome 39: 1035–1038. - PubMed
-
- Ahmed E. A., van der Vaart A., Barten A., Kal H. B., Chen J., et al. , 2007. Differences in DNA double strand breaks repair in male germ cell types: lessons learned from a differential expression of Mdc1 and 53BP1. DNA Repair (Amst.) 6: 1243–1254. - PubMed
-
- Ahmed E. A., Philippens M. E., Kal H. B., de Rooij D. G., de Boer P., 2010. Genetic probing of homologous recombination and non-homologous end joining during meiotic prophase in irradiated mouse spermatocytes. Mutat. Res. 688: 12–18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

