An Uncharacterized Apocarotenoid-Derived Signal Generated in ζ-Carotene Desaturase Mutants Regulates Leaf Development and the Expression of Chloroplast and Nuclear Genes in Arabidopsis
- PMID: 24907342
- PMCID: PMC4114949
- DOI: 10.1105/tpc.114.123349
An Uncharacterized Apocarotenoid-Derived Signal Generated in ζ-Carotene Desaturase Mutants Regulates Leaf Development and the Expression of Chloroplast and Nuclear Genes in Arabidopsis
Abstract
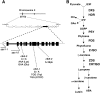
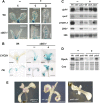
In addition to acting as photoprotective compounds, carotenoids also serve as precursors in the biosynthesis of several phytohormones and proposed regulatory signals. Here, we report a signaling process derived from carotenoids that regulates early chloroplast and leaf development. Biosynthesis of the signal depends on ζ-carotene desaturase activity encoded by the ζ-CAROTENE DESATURASE (ZDS)/CHLOROPLAST BIOGENESIS5 (CLB5) gene in Arabidopsis thaliana. Unlike other carotenoid-deficient plants, zds/clb5 mutant alleles display profound alterations in leaf morphology and cellular differentiation as well as altered expression of many plastid- and nucleus-encoded genes. The leaf developmental phenotypes and gene expression alterations of zds/clb5/spc1/pde181 plants are rescued by inhibitors or mutations of phytoene desaturase, demonstrating that phytofluene and/or ζ-carotene are substrates for an unidentified signaling molecule. Our work further demonstrates that this signal is an apocarotenoid whose synthesis requires the activity of the carotenoid cleavage dioxygenase CCD4.
© 2014 American Society of Plant Biologists. All rights reserved.
Figures





References
-
- Ahrazem O., Trapero A., Gómez M.D., Rubio-Moraga A., Gómez-Gómez L. (2010). Genomic analysis and gene structure of the plant carotenoid dioxygenase 4 family: A deeper study in Crocus sativus and its allies. Genomics 96: 239–250. - PubMed
-
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., Ghisla S., Bouwmeester H., Beyer P., Al-Babili S. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351. - PubMed
-
- Auldridge M.E., Block A., Vogel J.T., Dabney-Smith C., Mila I., Bouzayen M., Magallanes-Lundback M., DellaPenna D., McCarty D.R., Klee H.J. (2006a). Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 45: 982–993. - PubMed
-
- Auldridge M.E., McCarty D.R., Klee H.J. (2006b). Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 9: 315–321. - PubMed
-
- Barkan A., Goldschmidt-Clermont M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

