Genetics of ecological divergence during speciation
- PMID: 24909991
- PMCID: PMC4149549
- DOI: 10.1038/nature13301
Genetics of ecological divergence during speciation
Abstract
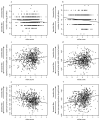
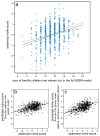
Ecological differences often evolve early in speciation as divergent natural selection drives adaptation to distinct ecological niches, leading ultimately to reproductive isolation. Although this process is a major generator of biodiversity, its genetic basis is still poorly understood. Here we investigate the genetic architecture of niche differentiation in a sympatric species pair of threespine stickleback fish by mapping the environment-dependent effects of phenotypic traits on hybrid feeding and performance under semi-natural conditions. We show that multiple, unlinked loci act largely additively to determine position along the major niche axis separating these recently diverged species. We also find that functional mismatch between phenotypic traits reduces the growth of some stickleback hybrids beyond that expected from an intermediate phenotype, suggesting a role for epistasis between the underlying genes. This functional mismatch might lead to hybrid incompatibilities that are analogous to those underlying intrinsic reproductive isolation but depend on the ecological context.
Figures







Comment in
-
Natural selection: it's a many-small world after all.Curr Biol. 2014 Oct 6;24(19):R959-62. doi: 10.1016/j.cub.2014.09.005. Curr Biol. 2014. PMID: 25291637
References
-
- Darwin C. The Origin of Species by Means of Natural Selection. John Murray; 1859.
-
- Schluter D. The Ecology of Adaptive Radiation. Oxford University Press; 2000.
-
- Coyne JA, Orr HA. Speciation. Sinauer Associates, Inc.; 2004.
-
- Nosil P. Ecological Speciation. Oxford University Press; 2012.
-
- Chase JM, Leibold MA. Ecological Niches: Linking Classical and Contemporary Approaches. The University of Chicago Press; 2003.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

