Fludarabine downregulates indoleamine 2,3-dioxygenase in tumors via a proteasome-mediated degradation mechanism
- PMID: 24911872
- PMCID: PMC4050125
- DOI: 10.1371/journal.pone.0099211
Fludarabine downregulates indoleamine 2,3-dioxygenase in tumors via a proteasome-mediated degradation mechanism
Abstract
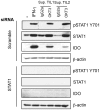
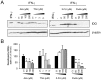
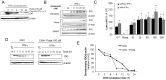
Indoleamine 2,3-dioxygenase (IDO) is found in multiple malignancies and exerts immunosuppressive effects that are central in protecting tumors from host T lymphocyte rejection. IDO is an enzyme involved in the catabolism of tryptophan resulting in inhibition of T lymphocyte function. While inhibition of IDO enzymatic activity results in tumor rejection, it is still unknown how we can directly target IDO expression within tumors using drugs. We have chosen to interfere with IDO expression by targeting the key-signaling event signal transducer and activator of transcription 1 (STAT1). We evaluated the efficacy of fludarabine, previously described to inhibit STAT1 phosphorylation. Interestingly, fludarabine was efficient in suppressing protein expression and consequently IDO activity in two different cell lines derived from breast cancer and melanoma when IDO was activated with interferon-gamma (IFN-γ) or supernatants prepared from activated T lymphocytes. However, fludarabine had no inhibitory effect on STAT1 phosphorylation. Other IFN-γ-responsive genes were only marginally inhibited by fludarabine. The level of IDO transcript was unaffected by this inhibitor, suggesting the involvement of post-transcriptional control. Strikingly, we have found that the inhibition of proteasome partially protected IDO from fludarabine-induced degradation, indicating that fludarabine induces IDO degradation through a proteasome-dependent pathway. Currently used in the clinic to treat some malignancies, fludarabine has the potential for use in the treatment of human tumors through induction of IDO degradation and consequently, for the promotion of T cell-mediated anti-tumor response.
Conflict of interest statement
Figures



References
-
- Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, et al. (1998) Prevention of allogeneic fetal rejection by tryptophan catabolism. Science 281: 1191–1193. - PubMed
-
- Godin-Ethier J, Hanafi LA, Piccirillo CA, Lapointe R (2011) Indoleamine 2,3-dioxygenase expression in human cancers: clinical and immunologic perspectives. Clin Cancer Res 17: 6985–6991. - PubMed
-
- Godin-Ethier J, Pelletier S, Hanafi LA, Gannon PO, Forget MA, et al. (2009) Human activated T lymphocytes modulate IDO expression in tumors through Th1/Th2 balance. J Immunol 183: 7752–7760. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

