Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo
- PMID: 24912195
- PMCID: PMC4060677
- DOI: 10.1073/pnas.1404616111
Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo
Abstract
Morphogenesis occurs in 3D space over time and is guided by coordinated gene expression programs. Here we use postembryonic development in Arabidopsis plants to investigate the genetic control of growth. We demonstrate that gene expression driving the production of the growth-stimulating hormone gibberellic acid and downstream growth factors is first induced within the radicle tip of the embryo. The center of cell expansion is, however, spatially displaced from the center of gene expression. Because the rapidly growing cells have very different geometry from that of those at the tip, we hypothesized that mechanical factors may contribute to this growth displacement. To this end we developed 3D finite-element method models of growing custom-designed digital embryos at cellular resolution. We used this framework to conceptualize how cell size, shape, and topology influence tissue growth and to explore the interplay of geometrical and genetic inputs into growth distribution. Our simulations showed that mechanical constraints are sufficient to explain the disconnect between the experimentally observed spatiotemporal patterns of gene expression and early postembryonic growth. The center of cell expansion is the position where genetic and mechanical facilitators of growth converge. We have thus uncovered a mechanism whereby 3D cellular geometry helps direct where genetically specified growth takes place.
Keywords: biomechanics; computational modeling; plant development; quantification; seed germination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

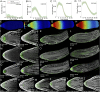
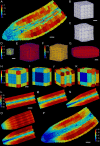

References
-
- Kuchen EE, et al. Generation of leaf shape through early patterns of growth and tissue polarity. Science. 2012;335(6072):1092–1096. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

