The Ctp type IVb pilus locus of Agrobacterium tumefaciens directs formation of the common pili and contributes to reversible surface attachment
- PMID: 24914181
- PMCID: PMC4135632
- DOI: 10.1128/JB.01670-14
The Ctp type IVb pilus locus of Agrobacterium tumefaciens directs formation of the common pili and contributes to reversible surface attachment
Abstract
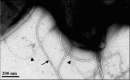
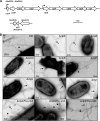
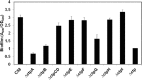
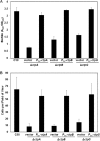
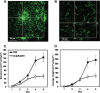
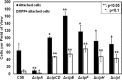
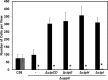
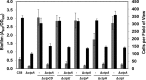
Agrobacterium tumefaciens can adhere to plant tissues and abiotic surfaces and forms biofilms. Cell surface appendages called pili play an important role in adhesion and biofilm formation in diverse bacterial systems. The A. tumefaciens C58 genome sequence revealed the presence of the ctpABCDEFGHI genes (cluster of type IV pili; Atu0216 to Atu0224), homologous to tad-type pilus systems from several bacteria, including Aggregatibacter actinomycetemcomitans and Caulobacter crescentus. These systems fall into the type IVb pilus group, which can function in bacterial adhesion. Transmission electron microscopy of A. tumefaciens revealed the presence of filaments, significantly thinner than flagella and often bundled, associated with cell surfaces and shed into the external milieu. In-frame deletion mutations of all of the ctp genes, with the exception of ctpF, resulted in nonpiliated derivatives. Mutations in ctpA (a pilin homologue), ctpB, and ctpG decreased early attachment and biofilm formation. The adherence of the ctpA mutant could be restored by ectopic expression of the paralogous pilA gene. The ΔctpA ΔpilA double pilin mutant displayed a diminished biovolume and lower biofilm height than the wild type under flowing conditions. Surprisingly, however, the ctpCD, ctpE, ctpF, ctpH, and ctpI mutants formed normal biofilms and showed enhanced reversible attachment. In-frame deletion of the ctpA pilin gene in the ctpCD, ctpE, ctpF, ctpH, and ctpI mutants caused the same attachment-deficient phenotype as the ctpA single mutant. Collectively, these findings indicate that the ctp locus is involved in pilus assembly and that nonpiliated mutants, which retain the CtpA pilin, are proficient in attachment and adherence.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Function and Regulation of Agrobacterium tumefaciens Cell Surface Structures that Promote Attachment.Curr Top Microbiol Immunol. 2018;418:143-184. doi: 10.1007/82_2018_96. Curr Top Microbiol Immunol. 2018. PMID: 29998422 Free PMC article. Review.
-
Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens.J Bacteriol. 2000 Jul;182(13):3705-16. doi: 10.1128/JB.182.13.3705-3716.2000. J Bacteriol. 2000. PMID: 10850985 Free PMC article.
-
VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus.J Bacteriol. 2001 Jun;183(12):3642-51. doi: 10.1128/JB.183.12.3642-3651.2001. J Bacteriol. 2001. PMID: 11371529 Free PMC article.
-
Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation.J Bacteriol. 2007 Nov;189(22):8005-14. doi: 10.1128/JB.00566-07. Epub 2007 Aug 31. J Bacteriol. 2007. PMID: 17766409 Free PMC article.
-
Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens.Curr Opin Microbiol. 2009 Dec;12(6):708-14. doi: 10.1016/j.mib.2009.09.014. Epub 2009 Oct 29. Curr Opin Microbiol. 2009. PMID: 19879182 Free PMC article. Review.
Cited by
-
Multiple Flagellin Proteins Have Distinct and Synergistic Roles in Agrobacterium tumefaciens Motility.J Bacteriol. 2018 Nov 6;200(23):e00327-18. doi: 10.1128/JB.00327-18. Print 2018 Dec 1. J Bacteriol. 2018. PMID: 30201783 Free PMC article.
-
Tad Pili Play a Dynamic Role in Caulobacter crescentus Surface Colonization.mBio. 2019 Jun 18;10(3):e01237-19. doi: 10.1128/mBio.01237-19. mBio. 2019. PMID: 31213565 Free PMC article.
-
Upstream CtrA-binding sites both induce and repress pilin gene expression in Caulobacter crescentus.BMC Genomics. 2024 Jul 19;25(1):703. doi: 10.1186/s12864-024-10533-6. BMC Genomics. 2024. PMID: 39030481 Free PMC article.
-
Glycoside Hydrolase Genes Are Required for Virulence of Agrobacterium tumefaciens on Bryophyllum daigremontiana and Tomato.Appl Environ Microbiol. 2019 Jul 18;85(15):e00603-19. doi: 10.1128/AEM.00603-19. Print 2019 Aug 1. Appl Environ Microbiol. 2019. PMID: 31126942 Free PMC article.
-
Mechanisms and regulation of surface interactions and biofilm formation in Agrobacterium.Front Plant Sci. 2014 May 6;5:176. doi: 10.3389/fpls.2014.00176. eCollection 2014. Front Plant Sci. 2014. PMID: 24834068 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

