The hnRNP-Q protein LIF2 participates in the plant immune response
- PMID: 24914891
- PMCID: PMC4051675
- DOI: 10.1371/journal.pone.0099343
The hnRNP-Q protein LIF2 participates in the plant immune response
Abstract
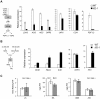
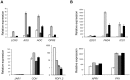
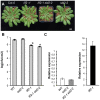
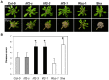
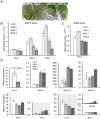
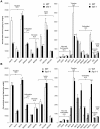
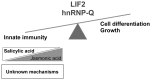
Eukaryotes have evolved complex defense pathways to combat invading pathogens. Here, we investigated the role of the Arabidopsis thaliana heterogeneous nuclear ribonucleoprotein (hnRNP-Q) LIF2 in the plant innate immune response. We show that LIF2 loss-of-function in A. thaliana leads to changes in the basal expression of the salicylic acid (SA)- and jasmonic acid (JA)- dependent defense marker genes PR1 and PDF1.2, respectively. Whereas the expression of genes involved in SA and JA biosynthesis and signaling was also affected in the lif2-1 mutant, no change in SA and JA hormonal contents was detected. In addition, the composition of glucosinolates, a class of defense-related secondary metabolites, was altered in the lif2-1 mutant in the absence of pathogen challenge. The lif2-1 mutant exhibited reduced susceptibility to the hemi-biotrophic pathogen Pseudomonas syringae and the necrotrophic ascomycete Botrytis cinerea. Furthermore, the lif2-1 sid2-2 double mutant was less susceptible than the wild type to P. syringae infection, suggesting that the lif2 response to pathogens was independent of SA accumulation. Together, our data suggest that lif2-1 exhibits a basal primed defense state, resulting from complex deregulation of gene expression, which leads to increased resistance to pathogens with various infection strategies. Therefore, LIF2 may function as a suppressor of cell-autonomous immunity. Similar to its human homolog, NSAP1/SYNCRIP, a trans-acting factor involved in both cellular processes and the viral life cycle, LIF2 may regulate the conflicting aspects of development and defense programs, suggesting that a conserved evolutionary trade-off between growth and defense pathways exists in eukaryotes.
Conflict of interest statement
Figures


Similar articles
-
Ethylene and jasmonic acid signaling affect the NPR1-independent expression of defense genes without impacting resistance to Pseudomonas syringae and Peronospora parasitica in the Arabidopsis ssi1 mutant.Mol Plant Microbe Interact. 2003 Jul;16(7):588-99. doi: 10.1094/MPMI.2003.16.7.588. Mol Plant Microbe Interact. 2003. PMID: 12848424
-
ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis.PLoS One. 2012;7(4):e35995. doi: 10.1371/journal.pone.0035995. Epub 2012 Apr 26. PLoS One. 2012. PMID: 22563431 Free PMC article.
-
Rhamnolipids elicit defense responses and induce disease resistance against biotrophic, hemibiotrophic, and necrotrophic pathogens that require different signaling pathways in Arabidopsis and highlight a central role for salicylic acid.Plant Physiol. 2012 Nov;160(3):1630-41. doi: 10.1104/pp.112.201913. Epub 2012 Sep 11. Plant Physiol. 2012. PMID: 22968829 Free PMC article.
-
The diversity of salicylic acid biosynthesis and defense signaling in plants: Knowledge gaps and future opportunities.Curr Opin Plant Biol. 2023 Apr;72:102349. doi: 10.1016/j.pbi.2023.102349. Epub 2023 Feb 24. Curr Opin Plant Biol. 2023. PMID: 36842224 Review.
-
Salicylic acid and jasmonic acid in elevated CO2-induced plant defense response to pathogens.J Plant Physiol. 2023 Jul;286:154019. doi: 10.1016/j.jplph.2023.154019. Epub 2023 May 20. J Plant Physiol. 2023. PMID: 37244001 Review.
Cited by
-
StMPK7 phosphorylates and stabilizes a potato RNA-binding protein StUBA2a/b to enhance plant defence responses.Hortic Res. 2022 Aug 24;9:uhac177. doi: 10.1093/hr/uhac177. eCollection 2022. Hortic Res. 2022. PMID: 36324643 Free PMC article.
-
Dissecting the Genetic Architecture of Morphological Traits in Sunflower (Helianthus annuus L.).Genes (Basel). 2024 Jul 19;15(7):950. doi: 10.3390/genes15070950. Genes (Basel). 2024. PMID: 39062729 Free PMC article.
-
The bile acid deoxycholate elicits defences in Arabidopsis and reduces bacterial infection.Mol Plant Pathol. 2017 May;18(4):540-554. doi: 10.1111/mpp.12416. Epub 2016 Jul 1. Mol Plant Pathol. 2017. PMID: 27085087 Free PMC article.
-
SYNCRIP facilitates porcine parvovirus viral DNA replication through the alternative splicing of NS1 mRNA to promote NS2 mRNA formation.Vet Res. 2021 May 25;52(1):73. doi: 10.1186/s13567-021-00938-6. Vet Res. 2021. PMID: 34034820 Free PMC article.
-
BdERECTA controls vasculature patterning and phloem-xylem organization in Brachypodium distachyon.BMC Plant Biol. 2021 Apr 23;21(1):196. doi: 10.1186/s12870-021-02970-2. BMC Plant Biol. 2021. PMID: 33892630 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

