Structural and functional analysis of the human spliceosomal DEAD-box helicase Prp28
- PMID: 24914973
- PMCID: PMC4051504
- DOI: 10.1107/S1399004714006439
Structural and functional analysis of the human spliceosomal DEAD-box helicase Prp28
Abstract
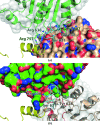
The DEAD-box protein Prp28 is essential for pre-mRNA splicing as it plays a key role in the formation of an active spliceosome. Prp28 participates in the release of the U1 snRNP from the 5'-splice site during association of the U5·U4/U6 tri-snRNP, which is a crucial step in the transition from a pre-catalytic spliceosome to an activated spliceosome. Here, it is demonstrated that the purified helicase domain of human Prp28 (hPrp28ΔN) binds ADP, whereas binding of ATP and ATPase activity could not be detected. ATP binding could not be observed for purified full-length hPrp28 either, but within an assembled spliceosomal complex hPrp28 gains ATP-binding activity. In order to understand the structural basis for the ATP-binding deficiency of isolated hPrp28, the crystal structure of hPrp28ΔN was determined at 2.0 Å resolution. In the crystal the helicase domain adopts a wide-open conformation, as the two RecA-like domains are extraordinarily displaced from the productive ATPase conformation. Binding of ATP is hindered by a closed conformation of the P-loop, which occupies the space required for the γ-phosphate of ATP.
Keywords: DEAD-box proteins; Prp28; helicase domain.
Figures

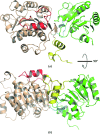


References
-
- Abrahams, J. P. & Leslie, A. G. W. (1996). Acta Cryst. D52, 30–42. - PubMed
-
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
-
- Benz, J., Trachsel, H. & Baumann, U. (1999). Structure, 7, 671–679. - PubMed
-
- Birdsall, B., King, R. W., Wheeler, M. R., Lewis, C. A. Jr, Goode, S. R., Dunlap, R. B. & Roberts, G. C. (1983). Anal. Biochem. 132, 353–361. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

