GlmU (N-acetylglucosamine-1-phosphate uridyltransferase) bound to three magnesium ions and ATP at the active site
- PMID: 24915076
- PMCID: PMC4051520
- DOI: 10.1107/S2053230X14008279
GlmU (N-acetylglucosamine-1-phosphate uridyltransferase) bound to three magnesium ions and ATP at the active site
Abstract
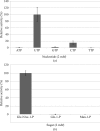
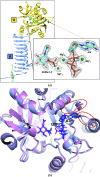
N-Acetylglucosamine-1-phosphate uridyltransferase (GlmU), a bifunctional enzyme exclusive to prokaryotes, belongs to the family of sugar nucleotidyltransferases (SNTs). The enzyme binds GlcNAc-1-P and UTP, and catalyzes a uridyltransfer reaction to synthesize UDP-GlcNAc, an important precursor for cell-wall biosynthesis. As many SNTs are known to utilize a broad range of substrates, substrate specificity in GlmU was probed using biochemical and structural studies. The enzymatic assays reported here demonstrate that GlmU is specific for its natural substrates UTP and GlcNAc-1-P. The crystal structure of GlmU bound to ATP and GlcNAc-1-P provides molecular details for the inability of the enzyme to utilize ATP for the nucleotidyltransfer reaction. ATP binding results in an inactive pre-catalytic enzyme-substrate complex, where it adopts an unusual conformation such that the reaction cannot be catalyzed; here, ATP is shown to be bound together with three Mg2+ ions. Overall, this structure represents the binding of an inhibitory molecule at the active site and can potentially be used to develop new inhibitors of the enzyme. Further, similar to DNA/RNA polymerases, GlmU was recently recognized to utilize two metal ions, MgA2+ and MgB2+, to catalyze the uridyltransfer reaction. Interestingly, displacement of MgB2+ from its usual catalytically competent position, as noted in the crystal structure of RNA polymerase in an inactive state, was considered to be a key factor inhibiting the reaction. Surprisingly, in the current structure of GlmU MgB2+ is similarly displaced; this raises the possibility that an analogous inhibitory mechanism may be operative in GlmU.
Keywords: ATP binding; enzyme inhibitor; substrate specificity; sugar nucleotidyltransferase.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

