Precision genome engineering and agriculture: opportunities and regulatory challenges
- PMID: 24915127
- PMCID: PMC4051594
- DOI: 10.1371/journal.pbio.1001877
Precision genome engineering and agriculture: opportunities and regulatory challenges
Abstract
Plant agriculture is poised at a technological inflection point. Recent advances in genome engineering make it possible to precisely alter DNA sequences in living cells, providing unprecedented control over a plant's genetic material. Potential future crops derived through genome engineering include those that better withstand pests, that have enhanced nutritional value, and that are able to grow on marginal lands. In many instances, crops with such traits will be created by altering only a few nucleotides among the billions that comprise plant genomes. As such, and with the appropriate regulatory structures in place, crops created through genome engineering might prove to be more acceptable to the public than plants that carry foreign DNA in their genomes. Public perception and the performance of the engineered crop varieties will determine the extent to which this powerful technology contributes towards securing the world's food supply.
Conflict of interest statement
D.F.V. is an inventor on several patents concerning TAL effector-mediated DNA modification and serves as Chief Science Officer for Cellectis Plant Sciences, a biotechnology company that uses sequence-specific nucleases to create new crop varieties.
Figures
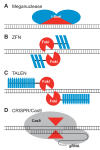

References
-
- Lippman ZB, Zamir D (2007) Heterosis: revisting the magic. Trends Genet 23: 60–66. - PubMed
-
- Clive J (2012) Global Status of Commercialized Biotech/GM Crops: 2012. ISAAA Brief No. 44. Ithaca (New York): ISAAA.
-
- Voytas DF (2013) Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 64: 327–350. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

