Characterization of the floral transcriptome of Moso bamboo (Phyllostachys edulis) at different flowering developmental stages by transcriptome sequencing and RNA-seq analysis
- PMID: 24915141
- PMCID: PMC4051636
- DOI: 10.1371/journal.pone.0098910
Characterization of the floral transcriptome of Moso bamboo (Phyllostachys edulis) at different flowering developmental stages by transcriptome sequencing and RNA-seq analysis
Abstract
Background: As an arborescent and perennial plant, Moso bamboo (Phyllostachys edulis (Carrière) J. Houzeau, synonym Phyllostachys heterocycla Carrière) is characterized by its infrequent sexual reproduction with flowering intervals ranging from several to more than a hundred years. However, little bamboo genomic research has been conducted on this due to a variety of reasons. Here, for the first time, we investigated the transcriptome of developing flowers in Moso bamboo by using high-throughput Illumina GAII sequencing and mapping short reads to the Moso bamboo genome and reference genes. We performed RNA-seq analysis on four important stages of flower development, and obtained extensive gene and transcript abundance data for the floral transcriptome of this key bamboo species.
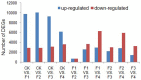
Results: We constructed a cDNA library using equal amounts of RNA from Moso bamboo leaf samples from non-flowering plants (CK) and mixed flower samples (F) of four flower development stages. We generated more than 67 million reads from each of the CK and F samples. About 70% of the reads could be uniquely mapped to the Moso bamboo genome and the reference genes. Genes detected at each stage were categorized to putative functional categories based on their expression patterns. The analysis of RNA-seq data of bamboo flowering tissues at different developmental stages reveals key gene expression properties during the flower development of bamboo.
Conclusion: We showed that a combination of transcriptome sequencing and RNA-seq analysis was a powerful approach to identifying candidate genes related to floral transition and flower development in bamboo species. The results give a better insight into the mechanisms of Moso bamboo flowering and ageing. This transcriptomic data also provides an important gene resource for improving breeding for Moso bamboo.
Conflict of interest statement
Figures




References
-
- Schuster SC (2008) Next-generation sequencing transforms today's biology. Nat Methods 5(1): 16–18 p. - PubMed
-
- Ansorge WJ (2009) Next-generation DNA sequencing techniques. N Biotechnol 25(4): 195–203. - PubMed
-
- Metzker ML (2010) Sequencing technologies-the next generation. Nat Rev Genet 11: 31–46. - PubMed
-
- Rosenkranz R, Borodina T, Lehrach H, Himmelbauer H (2008) Characterizing the mouse ES cell transcriptome with Illumina sequencing. Genomics 92: 187–194. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

