Similar patterns of clonally expanded somatic mtDNA mutations in the colon of heterozygous mtDNA mutator mice and ageing humans
- PMID: 24915468
- PMCID: PMC4141908
- DOI: 10.1016/j.mad.2014.06.003
Similar patterns of clonally expanded somatic mtDNA mutations in the colon of heterozygous mtDNA mutator mice and ageing humans
Abstract
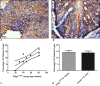
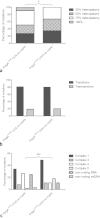
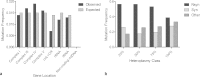
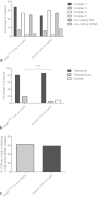
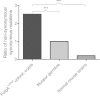
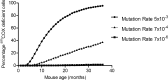
Clonally expanded mitochondrial DNA (mtDNA) mutations resulting in focal respiratory chain deficiency in individual cells are proposed to contribute to the ageing of human tissues that depend on adult stem cells for self-renewal; however, the consequences of these mutations remain unclear. A good animal model is required to investigate this further; but it is unknown whether mechanisms for clonal expansion of mtDNA mutations, and the mutational spectra, are similar between species. Here we show that mice, heterozygous for a mutation disrupting the proof-reading activity of mtDNA polymerase (PolgA(+/mut)) resulting in an increased mtDNA mutation rate, accumulate clonally expanded mtDNA point mutations in their colonic crypts with age. This results in focal respiratory chain deficiency, and by 81 weeks of age these animals exhibit a similar level and pattern of respiratory chain deficiency to 70-year-old human subjects. Furthermore, like in humans, the mtDNA mutation spectrum appears random and there is an absence of selective constraints. Computer simulations show that a random genetic drift model of mtDNA clonal expansion can accurately model the data from the colonic crypts of wild-type, PolgA(+/mut) animals, and humans, providing evidence for a similar mechanism for clonal expansion of mtDNA point mutations between these mice and humans.
Keywords: Ageing; Colon; Mitochondria; Mouse; MtDNA.
Copyright © 2014 The Authors. Published by Elsevier Ireland Ltd.. All rights reserved.
Figures

References
-
- Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., Schreier P.H., Smith A.J., Staden R., Young I.G. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
-
- Coller H.A., Khrapko K., Bodyak N.D., Nekhaeva E., Herrero-Jimenez P., Thilly W.G. High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat. Genet. 2001;28:147–150. - PubMed
-
- Coller H.A., Khrapko K., Herrero-Jimenez P., Vatland J.A., Li-Sucholeiki X.C., Thilly W.G. Clustering of mutant mitochondrial DNA copies suggests stem cells are common in human bronchial epithelium. Mutat. Res. 2005;578:256–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/H011471/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C008200/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 096919/Z/11/Z/WT_/Wellcome Trust/United Kingdom
- BB/F016980/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 096919/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

