The oxygen sensor MgFnr controls magnetite biomineralization by regulation of denitrification in Magnetospirillum gryphiswaldense
- PMID: 24915802
- PMCID: PMC4065386
- DOI: 10.1186/1471-2180-14-153
The oxygen sensor MgFnr controls magnetite biomineralization by regulation of denitrification in Magnetospirillum gryphiswaldense
Abstract
Background: Magnetotactic bacteria are capable of synthesizing magnetosomes only under oxygen-limited conditions. However, the mechanism of the aerobic repression on magnetite biomineralization has remained unknown. In Escherichia coli and other bacteria, Fnr (fumarate and nitrate reduction regulator) proteins are known to be involved in controlling the switch between microaerobic and aerobic metabolism. Here, we report on an Fnr-like protein (MgFnr) and its role in growth metabolism and magnetite biomineralization in the alphaproteobacterium Magnetospirillum gryphiswaldense.
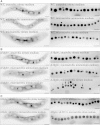
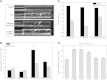
Results: Deletion of Mgfnr not only resulted in decreased N2 production due to reduced N2O reductase activity, but also impaired magnetite biomineralization under microaerobic conditions in the presence of nitrate. Overexpression of MgFnr in the WT also caused the synthesis of smaller magnetite particles under anaerobic and microaerobic conditions in the presence of nitrate. These data suggest that proper expression of MgFnr is required for WT-like magnetosome synthesis, which is regulated by oxygen. Analyses of transcriptional gusA reporter fusions revealed that besides showing similar properties to Fnr proteins reported in other bacteria, MgFnr is involved in the repression of the expression of denitrification genes nor and nosZ under aerobic conditions, possibly owing to several unique amino acid residues specific to MTB-Fnr.
Conclusions: We have identified and thoroughly characterized the first regulatory protein mediating denitrification growth and magnetite biomineralization in response to different oxygen conditions in a magnetotactic bacterium. Our findings reveal that the global oxygen regulator MgFnr is a genuine O2 sensor. It is involved in controlling expression of denitrification genes and thereby plays an indirect role in maintaining proper redox conditions required for magnetite biomineralization.
Figures


References
-
- Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacteriol. 2005;187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. - DOI - PMC - PubMed
-
- Lohsse A, Ullrich S, Katzmann E, Borg S, Wanner G, Richter M, Voigt B, Schweder T, Schüler D. Functional analysis of the magnetosome island in Magnetospirillum gryphiswaldense: the mamAB operon is sufficient for magnetite biomineralization. PLoS One. 2011;6:e25561. doi: 10.1371/journal.pone.0025561. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

