Evolutionary origins of human herpes simplex viruses 1 and 2
- PMID: 24916030
- PMCID: PMC4137711
- DOI: 10.1093/molbev/msu185
Evolutionary origins of human herpes simplex viruses 1 and 2
Abstract
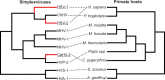
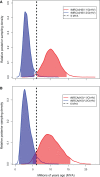
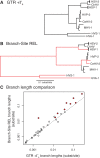
Herpesviruses have been infecting and codiverging with their vertebrate hosts for hundreds of millions of years. The primate simplex viruses exemplify this pattern of virus-host codivergence, at a minimum, as far back as the most recent common ancestor of New World monkeys, Old World monkeys, and apes. Humans are the only primate species known to be infected with two distinct herpes simplex viruses: HSV-1 and HSV-2. Human herpes simplex viruses are ubiquitous, with over two-thirds of the human population infected by at least one virus. Here, we investigated whether the additional human simplex virus is the result of ancient viral lineage duplication or cross-species transmission. We found that standard phylogenetic models of nucleotide substitution are inadequate for distinguishing among these competing hypotheses; the extent of synonymous substitutions causes a substantial underestimation of the lengths of some of the branches in the phylogeny, consistent with observations in other viruses (e.g., avian influenza, Ebola, and coronaviruses). To more accurately estimate ancient viral divergence times, we applied a branch-site random effects likelihood model of molecular evolution that allows the strength of natural selection to vary across both the viral phylogeny and the gene alignment. This selection-informed model favored a scenario in which HSV-1 is the result of ancient codivergence and HSV-2 arose from a cross-species transmission event from the ancestor of modern chimpanzees to an extinct Homo precursor of modern humans, around 1.6 Ma. These results provide a new framework for understanding human herpes simplex virus evolution and demonstrate the importance of using selection-informed models of sequence evolution when investigating viral origin hypotheses.
Keywords: co-divergence; cross-species transmission; homo; molecular clock; selection; zoonosis.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. - PubMed
-
- Anton SC. Natural history of Homo erectus. Am J Phys Anthropol. 2003;Suppl 37:126–170. - PubMed
-
- Brugha R, Keersmaekers K, Renton A, Meheus A. Genital herpes infection: a review. Int J Epidemiol. 1997;26:698–709. - PubMed
-
- Centers for Disease Control and Prevention. Epidemiologic notes and reports B-virus infection in Humans—Pensacola, Florida. Morb Mortal Wkly Rep. 1987;39:289–290. - PubMed

