Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing
- PMID: 24916110
- PMCID: PMC4134749
- DOI: 10.1161/CIRCRESAHA.115.304351
Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing
Abstract
Rationale: Individuals with naturally occurring loss-of-function proprotein convertase subtilisin/kexin type 9 (PCSK9) mutations experience reduced low-density lipoprotein cholesterol levels and protection against cardiovascular disease.
Objective: The goal of this study was to assess whether genome editing using a clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated system can efficiently introduce loss-of-function mutations into the endogenous PCSK9 gene in vivo.
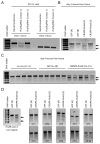
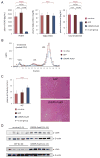
Methods and results: We used adenovirus to express CRISPR-associated 9 and a CRISPR guide RNA targeting Pcsk9 in mouse liver, where the gene is specifically expressed. We found that <3 to 4 days of administration of the virus, the mutagenesis rate of Pcsk9 in the liver was as high as >50%. This resulted in decreased plasma PCSK9 levels, increased hepatic low-density lipoprotein receptor levels, and decreased plasma cholesterol levels (by 35-40%). No off-target mutagenesis was detected in 10 selected sites.
Conclusions: Genome editing with the CRISPR-CRISPR-associated 9 system disrupts the Pcsk9 gene in vivo with high efficiency and reduces blood cholesterol levels in mice. This approach may have therapeutic potential for the prevention of cardiovascular disease in humans.
Keywords: coronary disease; genetic therapy; lipoproteins; molecular biology; prevention and control.
© 2014 American Heart Association, Inc.
Figures
Comment in
-
Peeking into a cool future: genome editing to delete PCSK9 and control hypercholesterolemia in a single shot.Circ Res. 2014 Aug 15;115(5):472-4. doi: 10.1161/CIRCRESAHA.114.304575. Circ Res. 2014. PMID: 25124321 Free PMC article. No abstract available.
-
One-shot, one cure with genome editing for dyslipidemia.Circ Cardiovasc Genet. 2014 Dec;7(6):967-8. doi: 10.1161/CIRCGENETICS.114.000958. Circ Cardiovasc Genet. 2014. PMID: 25516627 No abstract available.
References
-
- Abifadel M, Varret M, Rabès JP, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. - PubMed
-
- Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. - PubMed
-
- Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. - PubMed
-
- Hooper AJ, Marais AD, Tanyanyiwa DM, Burnett JR. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis. 2007;193:445–448. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

