Kidney cancer is characterized by aberrant methylation of tissue-specific enhancers that are prognostic for overall survival
- PMID: 24916699
- PMCID: PMC4424050
- DOI: 10.1158/1078-0432.CCR-14-0494
Kidney cancer is characterized by aberrant methylation of tissue-specific enhancers that are prognostic for overall survival
Abstract
Purpose: Even though recent studies have shown that genetic changes at enhancers can influence carcinogenesis, most methylomic studies have focused on changes at promoters. We used renal cell carcinoma (RCC), an incurable malignancy associated with mutations in epigenetic regulators, as a model to study genome-wide patterns of DNA methylation at a high resolution.
Experimental design: Analysis of cytosine methylation status of 1.3 million CpGs was determined by the HELP assay in RCC and healthy microdissected renal tubular controls.
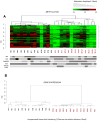
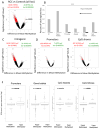
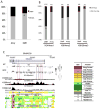
Results: We observed that the RCC samples were characterized by widespread hypermethylation that preferentially affected gene bodies. Aberrant methylation was particularly enriched in kidney-specific enhancer regions associated with H3K4Me1 marks. Various important underexpressed genes, such as SMAD6, were associated with aberrantly methylated, intronic enhancers, and these changes were validated in an independent cohort. MOTIF analysis of aberrantly hypermethylated regions revealed enrichment for binding sites of AP2a, AHR, HAIRY, ARNT, and HIF1 transcription factors, reflecting contributions of dysregulated hypoxia signaling pathways in RCC. The functional importance of this aberrant hypermethylation was demonstrated by selective sensitivity of RCC cells to low levels of decitabine. Most importantly, methylation of enhancers was predictive of adverse prognosis in 405 cases of RCC in multivariate analysis. In addition, parallel copy-number analysis from MspI representations demonstrated novel copy-number variations that were validated in an independent cohort of patients.
Conclusions: Our study is the first high-resolution methylome analysis of RCC, demonstrates that many kidney-specific enhancers are targeted by aberrant hypermethylation, and reveals the prognostic importance of these epigenetic changes in an independent cohort.
©2014 American Association for Cancer Research.
Figures



References
-
- Lamprecht B, Walter K, Kreher S, Kumar R, Hummel M, Lenze D, et al. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nature medicine. 2010;16:571–9. 1p following 9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

