Estimating quantitative genetic parameters in wild populations: a comparison of pedigree and genomic approaches
- PMID: 24917482
- PMCID: PMC4149785
- DOI: 10.1111/mec.12827
Estimating quantitative genetic parameters in wild populations: a comparison of pedigree and genomic approaches
Abstract
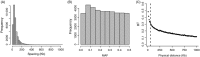
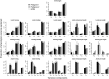
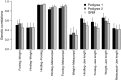
The estimation of quantitative genetic parameters in wild populations is generally limited by the accuracy and completeness of the available pedigree information. Using relatedness at genomewide markers can potentially remove this limitation and lead to less biased and more precise estimates. We estimated heritability, maternal genetic effects and genetic correlations for body size traits in an unmanaged long-term study population of Soay sheep on St Kilda using three increasingly complete and accurate estimates of relatedness: (i) Pedigree 1, using observation-derived maternal links and microsatellite-derived paternal links; (ii) Pedigree 2, using SNP-derived assignment of both maternity and paternity; and (iii) whole-genome relatedness at 37 037 autosomal SNPs. In initial analyses, heritability estimates were strikingly similar for all three methods, while standard errors were systematically lower in analyses based on Pedigree 2 and genomic relatedness. Genetic correlations were generally strong, differed little between the three estimates of relatedness and the standard errors declined only very slightly with improved relatedness information. When partitioning maternal effects into separate genetic and environmental components, maternal genetic effects found in juvenile traits increased substantially across the three relatedness estimates. Heritability declined compared to parallel models where only a maternal environment effect was fitted, suggesting that maternal genetic effects are confounded with direct genetic effects and that more accurate estimates of relatedness were better able to separate maternal genetic effects from direct genetic effects. We found that the heritability captured by SNP markers asymptoted at about half the SNPs available, suggesting that denser marker panels are not necessarily required for precise and unbiased heritability estimates. Finally, we present guidelines for the use of genomic relatedness in future quantitative genetics studies in natural populations.
Keywords: SNP genotyping; body size; genetic architecture; genomic relatedness; heritability; maternal genetic effect.
© 2014 The Authors. Molecular Ecology Published by John Wiley & Sons Ltd.
Figures

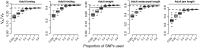
References
-
- Banos G, Wiggans GR, Powell RL. Impact of paternity errors in cow identification on genetic evaluations and international comparisons. Journal of Dairy Science. 2001;84:2523–2529. - PubMed
-
- Beraldi D, McRae AF, Gratten J, et al. Mapping quantitative trait loci underlying fitness-related traits in a free-living sheep population. Evolution. 2007;61:1403–1416. - PubMed
-
- Bijma P, Wade MJ. The joint effects of kin, multilevel selection and indirect genetic effects on response to genetic selection. Journal of Evolutionary Biology. 2008;21:1175–1188. - PubMed
-
- Blanckenhorn WU. The evolution of body size: What keeps organisms small? Quarterly Review of Biology. 2000;75:385–407. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

