Copy number variants and selective sweeps in natural populations of the house mouse (Mus musculus domesticus)
- PMID: 24917877
- PMCID: PMC4042557
- DOI: 10.3389/fgene.2014.00153
Copy number variants and selective sweeps in natural populations of the house mouse (Mus musculus domesticus)
Abstract
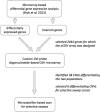
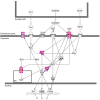
Copy-number variants (CNVs) may play an important role in early adaptations, potentially facilitating rapid divergence of populations. We describe an approach to study this question by investigating CNVs present in natural populations of mice in the early stages of divergence and their involvement in selective sweeps. We have analyzed individuals from two recently diverged natural populations of the house mouse (Mus musculus domesticus) from Germany and France using custom, high-density, comparative genome hybridization arrays (CGH) that covered almost 164 Mb and 2444 genes. One thousand eight hundred and sixty one of those genes we previously identified as differentially expressed between these populations, while the expression of the remaining genes was invariant. In total, we identified 1868 CNVs across all 10 samples, 200 bp to 600 kb in size and affecting 424 genic regions. Roughly two thirds of all CNVs found were deletions. We found no enrichment of CNVs among the differentially expressed genes between the populations compared to the invariant ones, nor any meaningful correlation between CNVs and gene expression changes. Among the CNV genes, we found cellular component gene ontology categories of the synapse overrepresented among all the 2444 genes tested. To investigate potential adaptive significance of the CNV regions, we selected six that showed large differences in frequency of CNVs between the two populations and analyzed variation in at least two microsatellites surrounding the loci in a sample of 46 unrelated animals from the same populations collected in field trappings. We identified two loci with large differences in microsatellite heterozygosity (Sfi1 and Glo1/Dnahc8 regions) and one locus with low variation across the populations (Cmah), thus suggesting that these genomic regions might have recently undergone selective sweeps. Interestingly, the Glo1 CNV has previously been implicated in anxiety-like behavior in mice, suggesting a differential evolution of a behavioral trait.
Keywords: CGH microarray; CNV; mice; natural populations; selective sweeps.
Figures


Similar articles
-
Divergence patterns of genic copy number variation in natural populations of the house mouse (Mus musculus domesticus) reveal three conserved genes with major population-specific expansions.Genome Res. 2015 Aug;25(8):1114-24. doi: 10.1101/gr.187187.114. Epub 2015 Jul 6. Genome Res. 2015. PMID: 26149421 Free PMC article.
-
An analysis of signatures of selective sweeps in natural populations of the house mouse.Mol Biol Evol. 2006 Apr;23(4):790-7. doi: 10.1093/molbev/msj096. Epub 2006 Jan 18. Mol Biol Evol. 2006. PMID: 16421176
-
Inter- and intra-breed genome-wide copy number diversity in a large cohort of European equine breeds.BMC Genomics. 2019 Oct 22;20(1):759. doi: 10.1186/s12864-019-6141-z. BMC Genomics. 2019. PMID: 31640551 Free PMC article.
-
Tissue-Specific eQTL in Zebrafish.Methods Mol Biol. 2020;2082:239-249. doi: 10.1007/978-1-0716-0026-9_17. Methods Mol Biol. 2020. PMID: 31849020
-
An evolving view of copy number variants.Curr Genet. 2019 Dec;65(6):1287-1295. doi: 10.1007/s00294-019-00980-0. Epub 2019 May 10. Curr Genet. 2019. PMID: 31076843 Review.
Cited by
-
The role of copy-number variation in the reinforcement of sexual isolation between the two European subspecies of the house mouse.Philos Trans R Soc Lond B Biol Sci. 2020 Aug 31;375(1806):20190540. doi: 10.1098/rstb.2019.0540. Epub 2020 Jul 13. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 32654648 Free PMC article.
-
A copy number variant is associated with a spectrum of pigmentation patterns in the rock pigeon (Columba livia).PLoS Genet. 2020 May 20;16(5):e1008274. doi: 10.1371/journal.pgen.1008274. eCollection 2020 May. PLoS Genet. 2020. PMID: 32433666 Free PMC article.
-
R2d2 Drives Selfish Sweeps in the House Mouse.Mol Biol Evol. 2016 Jun;33(6):1381-95. doi: 10.1093/molbev/msw036. Epub 2016 Feb 15. Mol Biol Evol. 2016. PMID: 26882987 Free PMC article.
-
Ecological and evolutionary implications of genomic structural variations.Front Genet. 2014 Sep 16;5:326. doi: 10.3389/fgene.2014.00326. eCollection 2014. Front Genet. 2014. PMID: 25278961 Free PMC article. No abstract available.
-
Divergence patterns of genic copy number variation in natural populations of the house mouse (Mus musculus domesticus) reveal three conserved genes with major population-specific expansions.Genome Res. 2015 Aug;25(8):1114-24. doi: 10.1101/gr.187187.114. Epub 2015 Jul 6. Genome Res. 2015. PMID: 26149421 Free PMC article.
References
-
- Cucchi T., Vigne J. D., Auffray J. (2005). First occurrence of the house mouse (Mus musculus domesticus Schwarz and Schwarz, 1943) in the Western Mediterranean: a zooarchaeological revision of subfossil occurrences. Biol. J. Linn. Soc. 84, 429–445 10.1111/j.1095-8312.2005.00445.x - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

