Genetic-based prediction of disease traits: prediction is very difficult, especially about the future
- PMID: 24917882
- PMCID: PMC4040440
- DOI: 10.3389/fgene.2014.00162
Genetic-based prediction of disease traits: prediction is very difficult, especially about the future
Abstract
Translation of results from genetic findings to inform medical practice is a highly anticipated goal of human genetics. The aim of this paper is to review and discuss the role of genetics in medically-relevant prediction. Germline genetics presages disease onset and therefore can contribute prognostic signals that augment laboratory tests and clinical features. As such, the impact of genetic-based predictive models on clinical decisions and therapy choice could be profound. However, given that (i) medical traits result from a complex interplay between genetic and environmental factors, (ii) the underlying genetic architectures for susceptibility to common diseases are not well-understood, and (iii) replicable susceptibility alleles, in combination, account for only a moderate amount of disease heritability, there are substantial challenges to constructing and implementing genetic risk prediction models with high utility. In spite of these challenges, concerted progress has continued in this area with an ongoing accumulation of studies that identify disease predisposing genotypes. Several statistical approaches with the aim of predicting disease have been published. Here we summarize the current state of disease susceptibility mapping and pharmacogenetics efforts for risk prediction, describe methods used to construct and evaluate genetic-based predictive models, and discuss applications.
Keywords: clinical utility; genetic risk; human genetics; predictive model; prognosis.
Figures

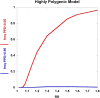

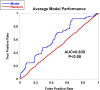
References
-
- Akaike H. (1974). A new look at the statistical model identification. Automatic Control IEEE Trans. 19, 716–723 10.1109/tac.1974.1100705 - DOI
-
- Aletaha D., Neogi T., Silman A. J., Funovits J., Felson D. T., Bingham C. O., 3rd, et al. (2010). 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann. Rheum. Dis. 69, 1580–1588 10.1136/ard.2010.138461 - DOI - PubMed
-
- Bao W., Hu F. B., Rong S., Rong Y., Bowers K., Schisterman E. F., et al. (2013). Predicting risk of type 2 diabetes mellitus with genetic risk models on the basis of established genome-wide association markers: a systematic review. Am. J. Epidemiol. 178, 1197–1207 10.1093/aje/kwt123 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

