Web services-based text-mining demonstrates broad impacts for interoperability and process simplification
- PMID: 24919658
- PMCID: PMC4207221
- DOI: 10.1093/database/bau050
Web services-based text-mining demonstrates broad impacts for interoperability and process simplification
Abstract
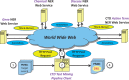
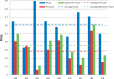
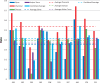
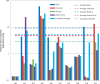
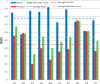
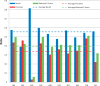
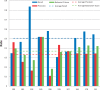
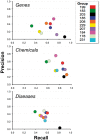
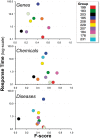
The Critical Assessment of Information Extraction systems in Biology (BioCreAtIvE) challenge evaluation tasks collectively represent a community-wide effort to evaluate a variety of text-mining and information extraction systems applied to the biological domain. The BioCreative IV Workshop included five independent subject areas, including Track 3, which focused on named-entity recognition (NER) for the Comparative Toxicogenomics Database (CTD; http://ctdbase.org). Previously, CTD had organized document ranking and NER-related tasks for the BioCreative Workshop 2012; a key finding of that effort was that interoperability and integration complexity were major impediments to the direct application of the systems to CTD's text-mining pipeline. This underscored a prevailing problem with software integration efforts. Major interoperability-related issues included lack of process modularity, operating system incompatibility, tool configuration complexity and lack of standardization of high-level inter-process communications. One approach to potentially mitigate interoperability and general integration issues is the use of Web services to abstract implementation details; rather than integrating NER tools directly, HTTP-based calls from CTD's asynchronous, batch-oriented text-mining pipeline could be made to remote NER Web services for recognition of specific biological terms using BioC (an emerging family of XML formats) for inter-process communications. To test this concept, participating groups developed Representational State Transfer /BioC-compliant Web services tailored to CTD's NER requirements. Participants were provided with a comprehensive set of training materials. CTD evaluated results obtained from the remote Web service-based URLs against a test data set of 510 manually curated scientific articles. Twelve groups participated in the challenge. Recall, precision, balanced F-scores and response times were calculated. Top balanced F-scores for gene, chemical and disease NER were 61, 74 and 51%, respectively. Response times ranged from fractions-of-a-second to over a minute per article. We present a description of the challenge and summary of results, demonstrating how curation groups can effectively use interoperable NER technologies to simplify text-mining pipeline implementation. Database URL: http://ctdbase.org/
© The Author(s) 2014. Published by Oxford University Press.
Figures

References
-
- Amberger J., Bocchini C., Hamosh A. (2011) A new face and new challenges for online mendelian inheritance in man (omim(r)). Hum Mutat., 32, 564–567 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

