Rewiring Host Lipid Metabolism by Large Viruses Determines the Fate of Emiliania huxleyi, a Bloom-Forming Alga in the Ocean
- PMID: 24920329
- PMCID: PMC4114960
- DOI: 10.1105/tpc.114.125641
Rewiring Host Lipid Metabolism by Large Viruses Determines the Fate of Emiliania huxleyi, a Bloom-Forming Alga in the Ocean
Abstract
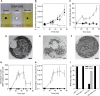
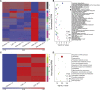
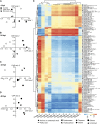
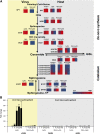
Marine viruses are major ecological and evolutionary drivers of microbial food webs regulating the fate of carbon in the ocean. We combined transcriptomic and metabolomic analyses to explore the cellular pathways mediating the interaction between the bloom-forming coccolithophore Emiliania huxleyi and its specific coccolithoviruses (E. huxleyi virus [EhV]). We show that EhV induces profound transcriptome remodeling targeted toward fatty acid synthesis to support viral assembly. A metabolic shift toward production of viral-derived sphingolipids was detected during infection and coincided with downregulation of host de novo sphingolipid genes and induction of the viral-encoded homologous pathway. The depletion of host-specific sterols during lytic infection and their detection in purified virions revealed their novel role in viral life cycle. We identify an essential function of the mevalonate-isoprenoid branch of sterol biosynthesis during infection and propose its downregulation as an antiviral mechanism. We demonstrate how viral replication depends on the hijacking of host lipid metabolism during the chemical "arms race" in the ocean.
© 2014 American Society of Plant Biologists. All rights reserved.
Figures



References
-
- Allen M.J., Martinez-Martinez J., Schroeder D.C., Somerfield P.J., Wilson W.H. (2007). Use of microarrays to assess viral diversity: from genotype to phenotype. Environ. Microbiol. 9: 971–982. - PubMed
-
- Anderson M.J., Willis T.J. (2003). Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84: 511–525.
LinkOut - more resources
Full Text Sources
Other Literature Sources

