RNA granule assembly and disassembly modulated by nuclear factor associated with double-stranded RNA 2 and nuclear factor 45
- PMID: 24920670
- PMCID: PMC4110319
- DOI: 10.1074/jbc.M114.556365
RNA granule assembly and disassembly modulated by nuclear factor associated with double-stranded RNA 2 and nuclear factor 45
Abstract
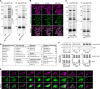
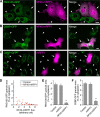
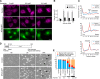
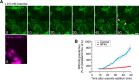
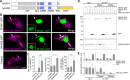
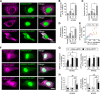
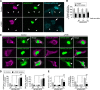
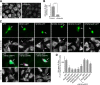
RNA granules are large messenger ribonucleoprotein complexes that regulate translation and mRNA translocation to control the timing and location of protein synthesis. The regulation of RNA granule assembly and disassembly is a structural basis of translational control, and its disorder is implicated in degenerative disease. Here, we used proteomic analysis to identify proteins associated with RNA granule protein 105 (RNG105)/caprin1, an RNA-binding protein in RNA granules. Among the identified proteins, we focused on nuclear factor (NF) 45 and its binding partner, nuclear factor associated with dsRNA 2 (NFAR2), and we demonstrated that NF45 promotes disassembly of RNA granules, whereas NFAR2 enhances the assembly of RNA granules in cultured cells. The GQSY domain of NFAR2 was required to associate with messenger ribonucleoprotein complexes containing RNG105/caprin1, and it was structurally and functionally related to the low complexity sequence domain of the fused in sarcoma protein, which drives the assembly of RNA granules. Another domain of NFAR2, the DZF domain, was dispensable for association with the RNG105 complex, but it was involved in positive and negative regulation of RNA granule assembly by being phosphorylated at double-stranded RNA-activated kinase sites and by association with NF45, respectively. These results suggest a novel molecular mechanism for the modulation of RNA granule assembly and disassembly by NFAR2, NF45, and phosphorylation at double-stranded RNA-activated kinase PKR sites.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

