The mechanism of dynein light chain LC8-mediated oligomerization of the Ana2 centriole duplication factor
- PMID: 24920673
- PMCID: PMC4110283
- DOI: 10.1074/jbc.M114.576041
The mechanism of dynein light chain LC8-mediated oligomerization of the Ana2 centriole duplication factor
Abstract
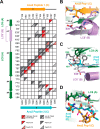
Centrioles play a key role in nucleating polarized microtubule networks. In actively dividing cells, centrioles establish the bipolar mitotic spindle and are essential for genomic stability. Drosophila anastral spindle-2 (Ana2) is a conserved centriole duplication factor. Although recent work has demonstrated that an Ana2-dynein light chain (LC8) centriolar complex is critical for proper spindle positioning in neuroblasts, how Ana2 and LC8 interact is yet to be established. Here we examine the Ana2-LC8 interaction and map two LC8-binding sites within the central region of Ana2, Ana2M (residues 156-251). Ana2 LC8-binding site 1 contains a signature TQT motif and robustly binds LC8 (KD of 1.1 μm), whereas site 2 contains a TQC motif and binds LC8 with lower affinity (KD of 13 μm). Both LC8-binding sites flank a predicted ~34-residue α-helix. We present two independent atomic structures of LC8 dimers in complex with Ana2 LC8-binding site 1 and site 2 peptides. The Ana2 peptides form β-strands that extend a central composite LC8 β-sandwich. LC8 recognizes the signature TQT motif in the first LC8 binding site of Ana2, forming extensive van der Waals contacts and hydrogen bonding with the peptide, whereas the Ana2 site 2 TQC motif forms a uniquely extended β-strand, not observed in other dynein light chain-target complexes. Size exclusion chromatography coupled with multiangle static light scattering demonstrates that LC8 dimers bind Ana2M sites and induce Ana2 tetramerization, yielding an Ana2M4-LC88 complex. LC8-mediated Ana2 oligomerization probably enhances Ana2 avidity for centriole-binding factors and may bridge multiple factors as required during spindle positioning and centriole biogenesis.
Keywords: Centriole; Centrosome; Cytoskeleton; Dynein; Isothermal Titration Calorimetry (ITC); Protein Complex; Protein Structure; Structural Biology.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



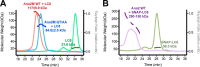
References
-
- Bornens M. (2012) The centrosome in cells and organisms. Science 335, 422–426 - PubMed
-
- Gönczy P. (2012) Towards a molecular architecture of centriole assembly. Nat. Rev. Mol. Cell Biol. 13, 425–435 - PubMed
-
- Guichard P., Hachet V., Majubu N., Neves A., Demurtas D., Olieric N., Fluckiger I., Yamada A., Kihara K., Nishida Y., Moriya S., Steinmetz M. O., Hongoh Y., Gönczy P. (2013) Native architecture of the centriole proximal region reveals features underlying its 9-fold radial symmetry. Curr. Biol. 23, 1620–1628 - PubMed
-
- Tsou M. F., Stearns T. (2006) Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947–951 - PubMed
-
- Pelletier L., O'Toole E., Schwager A., Hyman A. A., Müller-Reichert T. (2006) Centriole assembly in Caenorhabditis elegans. Nature 444, 619–623 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

