Genomic flexibility of human endogenous retrovirus type K
- PMID: 24920813
- PMCID: PMC4136327
- DOI: 10.1128/JVI.01147-14
Genomic flexibility of human endogenous retrovirus type K
Abstract
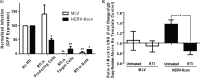
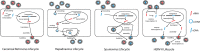
Human endogenous retrovirus type K (HERV-K) proviruses are scattered throughout the human genome, but as no infectious HERV-K virus has been detected to date, the mechanism by which these viruses replicated and populated the genome remains unresolved. Here, we provide evidence that, in addition to the RNA genomes that canonical retroviruses package, modern HERV-K viruses can contain reverse-transcribed DNA (RT-DNA) genomes. Indeed, reverse transcription of genomic HERV-K RNA into the DNA form is able to occur in three distinct times and locations: (i) in the virus-producing cell prior to viral release, yielding a DNA-containing extracellular virus particle similar to the spumaviruses; (ii) within the extracellular virus particle itself, transitioning from an RNA-containing particle to a DNA-containing particle; and (iii) after entry of the RNA-containing virus into the target cell, similar to canonical retroviruses, such as murine leukemia virus and HIV. Moreover, using a resuscitated HERV-K virus construct, we show that both viruses with RNA genomes and viruses with DNA genomes are capable of infecting target cells. This high level of genomic flexibility historically could have permitted these viruses to replicate in various host cell environments, potentially assisting in their many integration events and resulting in their high prevalence in the human genome. Moreover, the ability of modern HERV-K viruses to proceed through reverse transcription and package RT-DNA genomes suggests a higher level of replication competency than was previously understood, and it may be relevant in HERV-K-associated human diseases.
Importance: Retroviral elements comprise at least 8% of the human genome. Of all the endogenous retroviruses, HERV-K viruses are the most intact and biologically active. While a modern infectious HERV-K has yet to be found, HERV-K activation has been associated with cancers, autoimmune diseases, and HIV-1 infection. Thus, determining how this virus family became such a prevalent member of our genome and what it is capable of in its current form are of the utmost importance. Here, we provide evidence that HERV-K viruses currently found in the human genome are able to proceed through reverse transcription and historically utilized a life cycle with a surprising degree of genomic flexibility in which both RNA- and DNA-containing viruses were capable of mediating infection.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


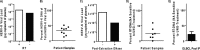

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

