ARH-seq: identification of differential splicing in RNA-seq data
- PMID: 24920826
- PMCID: PMC4132698
- DOI: 10.1093/nar/gku495
ARH-seq: identification of differential splicing in RNA-seq data
Abstract
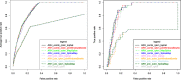
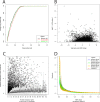
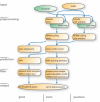
The computational prediction of alternative splicing from high-throughput sequencing data is inherently difficult and necessitates robust statistical measures because the differential splicing signal is overlaid by influencing factors such as gene expression differences and simultaneous expression of multiple isoforms amongst others. In this work we describe ARH-seq, a discovery tool for differential splicing in case-control studies that is based on the information-theoretic concept of entropy. ARH-seq works on high-throughput sequencing data and is an extension of the ARH method that was originally developed for exon microarrays. We show that the method has inherent features, such as independence of transcript exon number and independence of differential expression, what makes it particularly suited for detecting alternative splicing events from sequencing data. In order to test and validate our workflow we challenged it with publicly available sequencing data derived from human tissues and conducted a comparison with eight alternative computational methods. In order to judge the performance of the different methods we constructed a benchmark data set of true positive splicing events across different tissues agglomerated from public databases and show that ARH-seq is an accurate, computationally fast and high-performing method for detecting differential splicing events.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. - PubMed
-
- Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

