STI-GMaS: an open-source environment for simulation of sexually-transmitted infections
- PMID: 24923486
- PMCID: PMC4074422
- DOI: 10.1186/1752-0509-8-66
STI-GMaS: an open-source environment for simulation of sexually-transmitted infections
Abstract
Background: Sexually-transmitted pathogens often have severe reproductive health implications if treatment is delayed or absent, especially in females. The complex processes of disease progression, namely replication and ascension of the infection through the genital tract, span both extracellular and intracellular physiological scales, and in females can vary over the distinct phases of the menstrual cycle. The complexity of these processes, coupled with the common impossibility of obtaining comprehensive and sequential clinical data from individual human patients, makes mathematical and computational modelling valuable tools in developing our understanding of the infection, with a view to identifying new interventions. While many within-host models of sexually-transmitted infections (STIs) are available in existing literature, these models are difficult to deploy in clinical/experimental settings since simulations often require complex computational approaches.
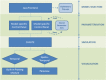
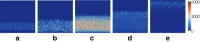
Results: We present STI-GMaS (Sexually-Transmitted Infections - Graphical Modelling and Simulation), an environment for simulation of STI models, with a view to stimulating the uptake of these models within the laboratory or clinic. The software currently focuses upon the representative case-study of Chlamydia trachomatis, the most common sexually-transmitted bacterial pathogen of humans. Here, we demonstrate the use of a hybrid PDE-cellular automata model for simulation of a hypothetical Chlamydia vaccination, demonstrating the effect of a vaccine-induced antibody in preventing the infection from ascending to above the cervix. This example illustrates the ease with which existing models can be adapted to describe new studies, and its careful parameterisation within STI-GMaS facilitates future tuning to experimental data as they arise.
Conclusions: STI-GMaS represents the first software designed explicitly for in-silico simulation of STI models by non-theoreticians, thus presenting a novel route to bridging the gap between computational and clinical/experimental disciplines. With the propensity for model reuse and extension, there is much scope within STI-GMaS to allow clinical and experimental studies to inform model inputs and drive future model development. Many of the modelling paradigms and software design principles deployed to date transfer readily to other STIs, both bacterial and viral; forthcoming releases of STI-GMaS will extend the software to incorporate a more diverse range of infections.
Figures





References
-
- Wilson D, Bowlin A, Bavoil P, Rank R. Ocular pathologic response elicited by Chlamydia organisms and the predictive value of quantitative modeling. J Infect Dis. 2009;199(12):1780–1789. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

