Transcription factors involved in retinogenesis are co-opted by the circadian clock following photoreceptor differentiation
- PMID: 24924194
- PMCID: PMC4146392
- DOI: 10.1242/dev.104380
Transcription factors involved in retinogenesis are co-opted by the circadian clock following photoreceptor differentiation
Abstract
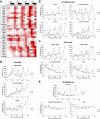
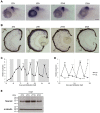
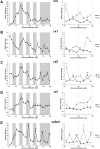
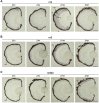
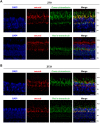
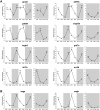
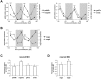
The circadian clock is known to regulate a wide range of physiological and cellular processes, yet remarkably little is known about its role during embryo development. Zebrafish offer a unique opportunity to explore this issue, not only because a great deal is known about key developmental events in this species, but also because the clock starts on the very first day of development. In this study, we identified numerous rhythmic genes in zebrafish larvae, including the key transcriptional regulators neurod and cdx1b, which are involved in neuronal and intestinal differentiation, respectively. Rhythmic expression of neurod and several additional transcription factors was only observed in the developing retina. Surprisingly, these rhythms in expression commenced at a stage of development after these transcription factors are known to have played their essential role in photoreceptor differentiation. Furthermore, this circadian regulation was maintained in adult retina. Thus, once mature photoreceptors are formed, multiple retinal transcription factors fall under circadian clock control, at which point they appear to play a new and important role in regulating rhythmic elements in the phototransduction pathway.
Keywords: Circadian clock; Photoreceptors; Retina; Transcription factors; Zebrafish; neurod.
© 2014. Published by The Company of Biologists Ltd.
Figures

References
-
- Blader P., Fischer N., Gradwohl G., Guillemot F., Strahle U. (1997). The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Development 124, 4557-4569 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

