Natural loss-of-function mutation of EDR1 conferring resistance to tomato powdery mildew in Arabidopsis thaliana accession C24
- PMID: 24925473
- PMCID: PMC6638503
- DOI: 10.1111/mpp.12165
Natural loss-of-function mutation of EDR1 conferring resistance to tomato powdery mildew in Arabidopsis thaliana accession C24
Abstract
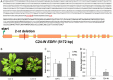
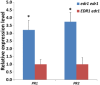
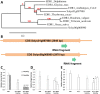
To screen for potentially novel types of resistance to tomato powdery mildew Oidium neolycopersici, a disease assay was performed on 123 Arabidopsis thaliana accessions. Forty accessions were fully resistant, and one, C24, was analysed in detail. By quantitative trait locus (QTL) analysis of an F2 population derived from C24 × Sha (susceptible accession), two QTLs associated with resistance were identified in C24. Fine mapping of QTL-1 on chromosome 1 delimited the region to an interval of 58 kb encompassing 15 candidate genes. One of these was Enhanced Disease Resistance 1 (EDR1). Evaluation of the previously obtained edr1 mutant of Arabidopsis accession Col-0, which was identified because of its resistance to powdery mildew Golovinomyces cichoracearum, showed that it also displayed resistance to O. neolycopersici. Sequencing of EDR1 in our C24 germplasm (referred to as C24-W) revealed two missing nucleotides in the second exon of EDR1 resulting in a premature stop codon. Remarkably, C24 obtained from other laboratories does not contain the EDR1 mutation. To verify the identity of C24-W, a DNA region containing a single nucleotide polymorphism (SNP) unique to C24 was sequenced showing that C24-W contains the C24-specific nucleotide. C24-W showed enhanced resistance to O. neolycopersici compared with C24 not containing the edr1 mutation. Furthermore, C24-W displayed a dwarf phenotype, which was not associated with the mutation in EDR1 and was not caused by the differential accumulation of pathogenesis-related genes. In conclusion, we identified a natural edr1 mutant in the background of C24.
Keywords: EDR1; Oidium neolycopersici; resistance; tomato.
© 2014 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures



References
-
- Alonso‐Blanco, C. and Koornneef, M. (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci. 5, 1360–1385. - PubMed
-
- Bai, Y. , Huang, C. , van der Hulst, R. , Meijer‐Dekens, F. , Bonnema, G. and Lindhout, P. (2003) QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorum G1.1601 co‐localize with two qualitative powdery mildew resistance genes. Mol. Plant–Microbe Interact. 16, 169–176. - PubMed
-
- Bai, Y. , van der Hulst, R. , Bonnema, G. , Marcel, T.L. , Meijer‐Dekens, F. , Niks, R. and Lindhout, P. (2005) Tomato defense to Oidium neolycopersici: dominant Ol genes confer isolate‐dependent resistance via a different mechanism than recessive ol‐2 . Mol. Plant–Microbe Interact. 18, 354–362. - PubMed
-
- Bai, Y. , Pavan, S. , Zheng, Z. , Zappel, N.F. , Reinstadler, A. , Lotti, C. , De Giovanni, C. , Ricciardi, L. , Lindhout, P. , Visser, R. , Theres, K. and Panstruga, R. (2008) Naturally occurring broad‐spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of mlo function. Mol. Plant–Microbe Interact. 21, 30–39. - PubMed
-
- Bechtold, U. , Lawson, T. , Mejia‐Carranza, J. , Meyer, R.C. , Brown, I.R. , Altmann, T. , Ton, J. and Mullineaux, P.M. (2010) Constitutive salicylic acid defences do not compromise seed yield, drought tolerance and water productivity in the Arabidopsis accession C24. Plant Cell Environ. 33, 1959–1973. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

