Targeting GTPases in Parkinson's disease: comparison to the historic path of kinase drug discovery and perspectives
- PMID: 24926233
- PMCID: PMC4046578
- DOI: 10.3389/fnmol.2014.00052
Targeting GTPases in Parkinson's disease: comparison to the historic path of kinase drug discovery and perspectives
Abstract
Neurological diseases have placed heavy social and financial burdens on modern society. As the life expectancy of humans is extended, neurological diseases, such as Parkinson's disease, have become increasingly common among senior populations. Although the enigmas of Parkinson's diseases await resolution, more vivid pictures on the cause, progression, and control of the illness are emerging after years of research. On the molecular level, GTPases are implicated in the etiology of Parkinson's disease and are rational pharmaceutical targets for their control. However, targeting individual GTPases, which belong to a superfamily of proteins containing multiple members with a conserved guanine nucleotide binding domain, has proven to be challenging. In contrast, pharmaceutical pursuit of inhibition of kinases, which constitute another superfamily of proteins with more than 500 members, has been fairly successful. We reviewed the breakthroughs in the history of kinase drug discovery to provide guidance for the GTPase field. We summarize recent progress made in the regulation of GTPase activity. We also present an efficient and cost effective approach to drug screening, which uses multiplex flow cytometry and mixture-based positional scanning libraries. These methods allow simultaneous measurements of both the activity and the selectivity of the screened library. Several GTPase activator clusters were identified which showed selectivity against different GTPase subfamilies. While the clusters need to be further deconvoluted to identify individual active compounds, the method described here and the structure information gathered create a foundation for further developments to build upon.
Keywords: GTPase; Parkinson’s; drug; kinase; multiplex.
Figures
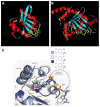
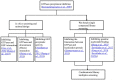
References
-
- Beilina A., Rudenko I. N., Kaganovich A., Civiero L., Chau H., Kalia S. K., et al. (2014). Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 111 2626–2631 10.1073/pnas.1318306111 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

