N-octanoyl dopamine treatment of endothelial cells induces the unfolded protein response and results in hypometabolism and tolerance to hypothermia
- PMID: 24926788
- PMCID: PMC4057113
- DOI: 10.1371/journal.pone.0099298
N-octanoyl dopamine treatment of endothelial cells induces the unfolded protein response and results in hypometabolism and tolerance to hypothermia
Abstract
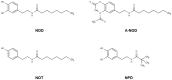
Aim: N-acyl dopamines (NADD) are gaining attention in the field of inflammatory and neurological disorders. Due to their hydrophobicity, NADD may have access to the endoplasmic reticulum (ER). We therefore investigated if NADD induce the unfolded protein response (UPR) and if this in turn influences cell behaviour.
Methods: Genome wide gene expression profiling, confirmatory qPCR and reporter assays were employed on human umbilical vein endothelial cells (HUVEC) to validate induction of UPR target genes and UPR sensor activation by N-octanoyl dopamine (NOD). Intracellular ATP, apoptosis and induction of thermotolerance were used as functional parameters to assess adaptation of HUVEC.
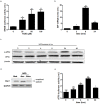
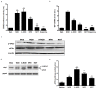
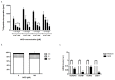
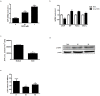
Results: NOD, but not dopamine dose dependently induces the UPR. This was also found for other synthetic NADD. Induction of the UPR was dependent on the redox activity of NADD and was not caused by selective activation of a particular UPR sensor. UPR induction did not result in cell apoptosis, yet NOD strongly impaired cell proliferation by attenuation of cells in the S-G2/M phase. Long-term treatment of HUVEC with low NOD concentration showed decreased intracellular ATP concentration paralleled with activation of AMPK. These cells were significantly more resistant to cold inflicted injury.
Conclusions: We provide for the first time evidence that NADD induce the UPR in vitro. It remains to be assessed if UPR induction is causally associated with hypometabolism and thermotolerance. Further pharmacokinetic studies are warranted to address if the NADD concentrations used in vitro can be obtained in vivo and if this in turn shows therapeutic efficacy.
Conflict of interest statement
Figures
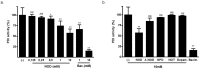

References
-
- Schroder M, Kaufman RJ (2005) The mammalian unfolded protein response. Annu Rev Biochem 74: 739–789. - PubMed
-
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529. - PubMed
-
- Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334: 1081–1086. - PubMed
-
- Kohno K (2010) Stress-sensing mechanisms in the unfolded protein response: similarities and differences between yeast and mammals. Journal of Biochemistry 147: 27–33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

