A new tool for monoclonal antibody analysis: application of IdeS proteolysis in IgG domain-specific characterization
- PMID: 24927271
- PMCID: PMC4171023
- DOI: 10.4161/mabs.28762
A new tool for monoclonal antibody analysis: application of IdeS proteolysis in IgG domain-specific characterization
Abstract
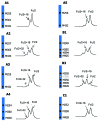
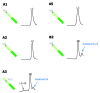
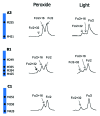
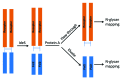
Monoclonal antibody (mAb) products are extraordinarily heterogeneous due to the presence of a variety of enzymatic and chemical modifications, such as deamidation, isomerization, oxidation, glycosylation, glycation, and terminal cyclization. The modifications in different domains of the antibody molecule can result in different biological consequences. Therefore, characterization and routine monitoring of domain-specific modifications are essential to ensure the quality of the therapeutic antibody products. For this purpose, a rapid and informative methodology was developed to examine the heterogeneity of individual domains in mAb products. A recently discovered endopeptidase, IdeS, cleaves heavy chains below the hinge region, producing F(ab') 2 and Fc fragments. Following reduction of disulfide bonds, three antibody domains (LC, Fd, and Fc/2) can be released for further characterization. Subsequent analyses by liquid chromatography/mass spectrometry, capillary isoelectric focusing, and glycan mapping enable domain-specific profiling of oxidation, charge heterogeneity, and glycoform distribution. When coupled with reversed phase chromatography, the unique chromatographic profile of each molecule offers a simple strategy for an identity test, which is an important formal test for biopharmaceutical quality control purposes. This methodology is demonstrated for a number of IgGs of different subclasses (IgG1, IgG2, IgG4), as well as an Fc fusion protein. The presented technique provides a convenient platform approach for scientific and formal therapeutic mAb product characterization. It can also be applied in regulated drug substance batch release and stability testing of antibody and Fc fusion protein products, in particular for identity and routine monitoring of domain-specific modifications.
Keywords: Fc fusion protein; IdeS; IgG; batch release; charge variant; domain-specific modification; formal identity; glycosylation; methionine oxidation; monoclonal antibody; stability testing.
Figures










References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
