Highly parallel characterization of IgG Fc binding interactions
- PMID: 24927273
- PMCID: PMC4171026
- DOI: 10.4161/mabs.28808
Highly parallel characterization of IgG Fc binding interactions
Abstract
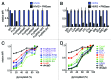
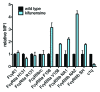
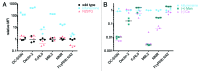
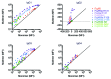
Because the variable ability of the antibody constant (Fc) domain to recruit innate immune effector cells and complement is a major factor in antibody activity in vivo, convenient means of assessing these binding interactions is of high relevance to the development of enhanced antibody therapeutics, and to understanding the protective or pathogenic antibody response to infection, vaccination, and self. Here, we describe a highly parallel microsphere assay to rapidly assess the ability of antibodies to bind to a suite of antibody receptors. Fc and glycan binding proteins such as FcγR and lectins were conjugated to coded microspheres and the ability of antibodies to interact with these receptors was quantified. We demonstrate qualitative and quantitative assessment of binding preferences and affinities across IgG subclasses, Fc domain point mutants, and antibodies with variant glycosylation. This method can serve as a rapid proxy for biophysical methods that require substantial sample quantities, high-end instrumentation, and serial analysis across multiple binding interactions, thereby offering a useful means to characterize monoclonal antibodies, clinical antibody samples, and antibody mimics, or alternatively, to investigate the binding preferences of candidate Fc receptors.
Keywords: Fc domain; Fcγ receptor; IgG; antibody; glycosylation; lectin; luminex; multiplex.
Figures




References
-
- Jefferis R, Pound J, Lund J, Goodall M. Effector mechanisms activated by human IgG subclass antibodies: clinical and molecular aspects. Review article. Ann Biol Clin (Paris) 1994;52:57–65. - PubMed
-
- Teeling JL, Jansen-Hendriks T, Kuijpers TW, de Haas M, van de Winkel JG, Hack CE, Bleeker WK. Therapeutic efficacy of intravenous immunoglobulin preparations depends on the immunoglobulin G dimers: studies in experimental immune thrombocytopenia. Blood. 2001;98:1095–9. doi: 10.1182/blood.V98.4.1095. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
