Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes
- PMID: 24927540
- PMCID: PMC4078802
- DOI: 10.1073/pnas.1401734111
Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes
Abstract
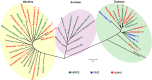
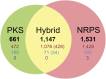
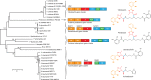
Nonribosomal peptides and polyketides are a diverse group of natural products with complex chemical structures and enormous pharmaceutical potential. They are synthesized on modular nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) enzyme complexes by a conserved thiotemplate mechanism. Here, we report the widespread occurrence of NRPS and PKS genetic machinery across the three domains of life with the discovery of 3,339 gene clusters from 991 organisms, by examining a total of 2,699 genomes. These gene clusters display extraordinarily diverse organizations, and a total of 1,147 hybrid NRPS/PKS clusters were found. Surprisingly, 10% of bacterial gene clusters lacked modular organization, and instead catalytic domains were mostly encoded as separate proteins. The finding of common occurrence of nonmodular NRPS differs substantially from the current classification. Sequence analysis indicates that the evolution of NRPS machineries was driven by a combination of common descent and horizontal gene transfer. We identified related siderophore NRPS gene clusters that encoded modular and nonmodular NRPS enzymes organized in a gradient. A higher frequency of the NRPS and PKS gene clusters was detected from bacteria compared with archaea or eukarya. They commonly occurred in the phyla of Proteobacteria, Actinobacteria, Firmicutes, and Cyanobacteria in bacteria and the phylum of Ascomycota in fungi. The majority of these NRPS and PKS gene clusters have unknown end products highlighting the power of genome mining in identifying novel genetic machinery for the biosynthesis of secondary metabolites.
Keywords: bioactive compound; biosynthetic gene cluster; data mining; distribution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Comment in
-
Comments on the distribution and phylogeny of type I polyketide synthases and nonribosomal peptide synthetases in eukaryotes.Proc Natl Acad Sci U S A. 2014 Sep 23;111(38):E3946. doi: 10.1073/pnas.1412766111. Epub 2014 Sep 2. Proc Natl Acad Sci U S A. 2014. PMID: 25197098 Free PMC article. No abstract available.
-
Reply to Sasso et al.: Distribution and phylogeny of nonribosomal peptide and polyketide biosynthetic pathways in eukaryotes.Proc Natl Acad Sci U S A. 2014 Sep 23;111(38):E3947. doi: 10.1073/pnas.1413343111. Epub 2014 Sep 2. Proc Natl Acad Sci U S A. 2014. PMID: 25199204 Free PMC article. No abstract available.
References
-
- Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: Combinations, permutations, and mutations. Science. 1998;282(5386):63–68. - PubMed
-
- Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides1. Annu Rev Microbiol. 2004;58:453–488. - PubMed
-
- Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbiol. 2005;3(12):925–936. - PubMed
-
- Kopp F, Marahiel MA. Where chemistry meets biology: the chemoenzymatic synthesis of nonribosomal peptides and polyketides. Curr Opin Biotechnol. 2007;18(6):513–520. - PubMed
-
- Walsh CT. The chemical versatility of natural-product assembly lines. Acc Chem Res. 2008;41(1):4–10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

