Self-processing of a barley subtilase expressed in E. coli
- PMID: 24927642
- PMCID: PMC4148201
- DOI: 10.1016/j.pep.2014.05.014
Self-processing of a barley subtilase expressed in E. coli
Abstract
The barley protease BAJ93208 belongs to the subtilase family of serine proteases. We have expressed BAJ93208 in the cytoplasm of the Escherichiacoli strain SHuffle C3030 using a rhamnose-inducible promoter. The expression construct included a (His)6-tag at the N-terminus and a strep-tag at the C-terminus. Western blot analysis revealed that the protein was processed at the N- and C-terminus. To exclude that this processing was due to contaminating E. coli proteases, a mutated BAJ93208 protease was constructed. This inactive mutant was not processed, demonstrating that the processing was an autocatalytic process. To define the exact cleavage sites mass spectrometry was used which detected four differently processed versions of the protease. At the N-terminus, the self-processing removed the internal inhibitor and an additional 19 amino acids. At the C-terminus there was a cleavage site after Ala(765) which also removed the strep-tag. This explained the inability to detect the purified (His)6-BAJ93208-strep protease with an anti-strep-tag antibody. Finally, an additional alanine was removed either at the N-terminus (Ala(119)) or at the C-terminus (Ala(764)).
Keywords: Proprotein; Subtilase; Thionin.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
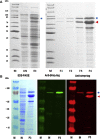

 indicates possible cleavage site which would remove one alanine.
indicates possible cleavage site which would remove one alanine.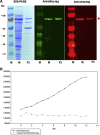
References
-
- Smith E.L. The complete amino acid sequence of two types of subtilisin, BPN’ and Carlsberg. J. Biol. Chem. 1966;241(24):5974–5976. - PubMed
-
- Julius D. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984;37(3):1075–1089. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

