Cortical and Clonal Contribution of Tbr2 Expressing Progenitors in the Developing Mouse Brain
- PMID: 24927931
- PMCID: PMC4585488
- DOI: 10.1093/cercor/bhu125
Cortical and Clonal Contribution of Tbr2 Expressing Progenitors in the Developing Mouse Brain
Abstract
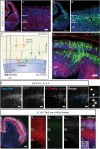
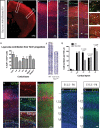
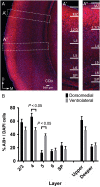
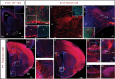
The individual contribution of different progenitor subtypes towards the mature rodent cerebral cortex is not fully understood. Intermediate progenitor cells (IPCs) are key to understanding the regulation of neuronal number during cortical development and evolution, yet their exact contribution is much debated. Intermediate progenitors in the cortical subventricular zone are defined by expression of T-box brain-2 (Tbr2). In this study we demonstrate by using the Tbr2(Cre) mouse line and state-of-the-art cell lineage labeling techniques, that IPC derived cells contribute substantial proportions 67.5% of glutamatergic but not GABAergic or astrocytic cells to all cortical layers including the earliest generated subplate zone. We also describe the laminar dispersion of clonally derived cells from IPCs using a recently described clonal analysis tool (CLoNe) and show that pair-generated cells in different layers cluster closer (142.1 ± 76.8 μm) than unrelated cells (294.9 ± 105.4 μm). The clonal dispersion from individual Tbr2 positive intermediate progenitors contributes to increasing the cortical surface. Our study also describes extracortical contributions from Tbr2+ progenitors to the lateral olfactory tract and ventromedial hypothalamic nucleus.
Keywords: CLoNe; Cerebral cortex; clonal analysis; fate-mapping; intermediate progenitors; subventricular zone.
© The Author 2014. Published by Oxford University Press.
Figures


References
-
- Arora S, Acharya J, Verma A, Panigrahi PK. 2008. Multilevel thresholding for image segmentation through a fast statistical recursive algorithm. Pattern Recognit Lett. 29:119–125.
-
- Baala L, Briault S, Etchevers HC, Laumonnier F, Natiq A, Amiel J, Boddaert N, Picard C, Sbiti A, Asermouh A, et al. 2007. Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat Genet. 39:454–456. - PubMed
-
- Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Ménard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, et al. 2013. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 80:442–457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

