Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4
- PMID: 24928045
- PMCID: PMC7111329
- DOI: 10.1016/j.virol.2014.04.027
Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4
Abstract
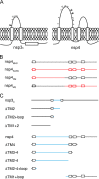
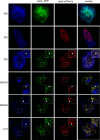
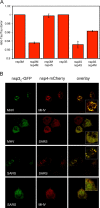
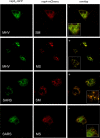
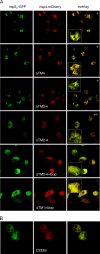
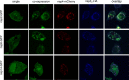
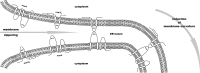
Coronaviruses replicate their genomes in association with rearranged cellular membranes. The coronavirus nonstructural integral membrane proteins (nsps) 3, 4 and 6, are key players in the formation of the rearranged membranes. Previously, we demonstrated that nsp3 and nsp4 interact and that their co-expression results in the relocalization of these proteins from the endoplasmic reticulum (ER) into discrete perinuclear foci. We now show that these foci correspond to areas of rearranged ER-derived membranes, which display increased membrane curvature. These structures, which were able to recruit other nsps, were only detected when nsp3 and nsp4 were derived from the same coronavirus species. We propose, based on the analysis of a large number of nsp3 and nsp4 mutants, that interaction between the large luminal loops of these proteins drives the formation of membrane rearrangements, onto which the coronavirus replication-transcription complexes assemble in infected cells.
Keywords: Coronavirus; Membrane rearrangements; Nsps; Replication–transcription complex; Transmembrane proteins.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication.mBio. 2017 Nov 21;8(6):e01658-17. doi: 10.1128/mBio.01658-17. mBio. 2017. PMID: 29162711 Free PMC article.
-
Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning.J Virol. 2008 Dec;82(24):12392-405. doi: 10.1128/JVI.01219-08. Epub 2008 Oct 8. J Virol. 2008. PMID: 18842706 Free PMC article.
-
Mutations across murine hepatitis virus nsp4 alter virus fitness and membrane modifications.J Virol. 2015 Feb;89(4):2080-9. doi: 10.1128/JVI.02776-14. Epub 2014 Dec 3. J Virol. 2015. PMID: 25473044 Free PMC article.
-
Biogenesis and architecture of arterivirus replication organelles.Virus Res. 2016 Jul 15;220:70-90. doi: 10.1016/j.virusres.2016.04.001. Epub 2016 Apr 9. Virus Res. 2016. PMID: 27071852 Free PMC article. Review.
-
Mechanism, structural and functional insights into nidovirus-induced double-membrane vesicles.Front Immunol. 2024 Jun 11;15:1340332. doi: 10.3389/fimmu.2024.1340332. eCollection 2024. Front Immunol. 2024. PMID: 38919631 Free PMC article. Review.
Cited by
-
Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles.Antiviral Res. 2016 Nov;135:97-107. doi: 10.1016/j.antiviral.2016.10.005. Epub 2016 Oct 13. Antiviral Res. 2016. PMID: 27743916 Free PMC article. Review.
-
Intragenomic rearrangements involving 5'-untranslated region segments in SARS-CoV-2, other betacoronaviruses, and alphacoronaviruses.Virol J. 2023 Feb 25;20(1):36. doi: 10.1186/s12985-023-01998-0. Virol J. 2023. PMID: 36829234 Free PMC article.
-
Renal Considerations in COVID-19: Biology, Pathology, and Pathophysiology.ASAIO J. 2021 Oct 1;67(10):1087-1096. doi: 10.1097/MAT.0000000000001530. ASAIO J. 2021. PMID: 34191753 Free PMC article.
-
A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis.PLoS Biol. 2020 Jun 8;18(6):e3000715. doi: 10.1371/journal.pbio.3000715. eCollection 2020 Jun. PLoS Biol. 2020. PMID: 32511245 Free PMC article.
-
SARS-CoV-2 variant-specific differences in inhibiting the effects of the PKR-activated integrated stress response.Virus Res. 2024 Jan 2;339:199271. doi: 10.1016/j.virusres.2023.199271. Epub 2023 Nov 28. Virus Res. 2024. PMID: 37979658 Free PMC article.
References
-
- Bernasconi R., Galli C., Noack J., Bianchi S., de Haan C.A., Reggiori F., Molinari M. Role of the SEL1L:LC3-I complex as an ERAD tuning receptor in the mammalian ER. Mol. Cell. 2012;46:809–819. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

