Norovirus translation requires an interaction between the C Terminus of the genome-linked viral protein VPg and eukaryotic translation initiation factor 4G
- PMID: 24928504
- PMCID: PMC4118132
- DOI: 10.1074/jbc.M114.550657
Norovirus translation requires an interaction between the C Terminus of the genome-linked viral protein VPg and eukaryotic translation initiation factor 4G
Abstract
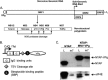
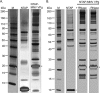
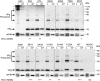
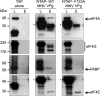
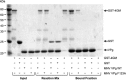
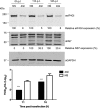
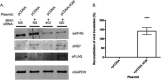
Viruses have evolved a variety of mechanisms to usurp the host cell translation machinery to enable translation of the viral genome in the presence of high levels of cellular mRNAs. Noroviruses, a major cause of gastroenteritis in man, have evolved a mechanism that relies on the interaction of translation initiation factors with the virus-encoded VPg protein covalently linked to the 5' end of the viral RNA. To further characterize this novel mechanism of translation initiation, we have used proteomics to identify the components of the norovirus translation initiation factor complex. This approach revealed that VPg binds directly to the eIF4F complex, with a high affinity interaction occurring between VPg and eIF4G. Mutational analyses indicated that the C-terminal region of VPg is important for the VPg-eIF4G interaction; viruses with mutations that alter or disrupt this interaction are debilitated or non-viable. Our results shed new light on the unusual mechanisms of protein-directed translation initiation.
Keywords: Eukaryotic Translation Initiation Factor 4E (eIF4E); Norovirus; Plus-stranded RNA Virus; Translation; Translation Initiation Factor; VPg; Virus; eIF4G.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Translational Control during Calicivirus Infection.Viruses. 2016 Apr 20;8(4):104. doi: 10.3390/v8040104. Viruses. 2016. PMID: 27104553 Free PMC article. Review.
-
A Conserved Interaction between a C-Terminal Motif in Norovirus VPg and the HEAT-1 Domain of eIF4G Is Essential for Translation Initiation.PLoS Pathog. 2016 Jan 6;12(1):e1005379. doi: 10.1371/journal.ppat.1005379. eCollection 2016 Jan. PLoS Pathog. 2016. PMID: 26734730 Free PMC article.
-
Sapovirus translation requires an interaction between VPg and the cap binding protein eIF4E.J Virol. 2014 Nov;88(21):12213-21. doi: 10.1128/JVI.01650-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142584 Free PMC article.
-
VPg of murine norovirus binds translation initiation factors in infected cells.Virol J. 2006 May 23;3:33. doi: 10.1186/1743-422X-3-33. Virol J. 2006. PMID: 16719923 Free PMC article.
-
Manipulation of the host translation initiation complex eIF4F by DNA viruses.Biochem Soc Trans. 2010 Dec;38(6):1511-6. doi: 10.1042/BST0381511. Biochem Soc Trans. 2010. PMID: 21118117 Review.
Cited by
-
Cell Cycle Arrest is a Conserved Function of Norovirus VPg Proteins.Viruses. 2019 Mar 4;11(3):217. doi: 10.3390/v11030217. Viruses. 2019. PMID: 30836641 Free PMC article.
-
Non-canonical Translation in Plant RNA Viruses.Front Plant Sci. 2017 Apr 6;8:494. doi: 10.3389/fpls.2017.00494. eCollection 2017. Front Plant Sci. 2017. PMID: 28428795 Free PMC article. Review.
-
Insight Into the Interaction Between RNA Polymerase and VPg for Murine Norovirus Replication.Front Microbiol. 2018 Jul 3;9:1466. doi: 10.3389/fmicb.2018.01466. eCollection 2018. Front Microbiol. 2018. PMID: 30038601 Free PMC article.
-
Translational Control during Calicivirus Infection.Viruses. 2016 Apr 20;8(4):104. doi: 10.3390/v8040104. Viruses. 2016. PMID: 27104553 Free PMC article. Review.
-
Interferon responses to norovirus infections: current and future perspectives.J Gen Virol. 2021 Oct;102(10):001660. doi: 10.1099/jgv.0.001660. J Gen Virol. 2021. PMID: 34698626 Free PMC article. Review.
References
-
- Lamphear B. J., Yan R., Yang F., Waters D., Liebig H. D., Klump H., Kuechler E., Skern T., Rhoads R. E. (1993) Mapping the cleavage site in protein synthesis initiation factor eIF-4γ of the 2A proteases from human coxsackievirus and rhinovirus. J. Biol. Chem. 268, 19200–19203 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/J001708/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I012303/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I01232X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 097997/WT_/Wellcome Trust/United Kingdom
- WT097997MA/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

