A nuclear ubiquitin-proteasome pathway targets the inner nuclear membrane protein Asi2 for degradation
- PMID: 24928896
- PMCID: PMC4333764
- DOI: 10.1242/jcs.153163
A nuclear ubiquitin-proteasome pathway targets the inner nuclear membrane protein Asi2 for degradation
Abstract
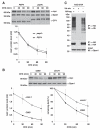
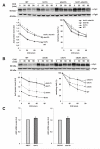
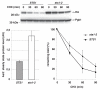
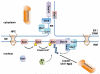
The nuclear envelope consists of inner and outer nuclear membranes. Whereas the outer membrane is an extension of the endoplasmic reticulum, the inner nuclear membrane (INM) represents a unique membranous environment containing specific proteins. The mechanisms of integral INM protein degradation are unknown. Here, we investigated the turnover of Asi2, an integral INM protein in Saccharomyces cerevisiae. We report that Asi2 is degraded by the proteasome independently of the vacuole and that it exhibited a half-life of ∼45 min. Asi2 exhibits enhanced stability in mutants lacking the E2 ubiquitin conjugating enzymes Ubc6 or Ubc7, or the E3 ubiquitin ligase Doa10. Consistent with these data, Asi2 is post-translationally modified by poly-ubiquitylation in a Ubc7- and Doa10-dependent manner. Importantly Asi2 degradation is significantly reduced in a sts1-2 mutant that fails to accumulate proteasomes in the nucleus, indicating that Asi2 is degraded in the nucleus. Our results reveal a molecular pathway that affects the stability of integral proteins of the inner nuclear membrane and indicate that Asi2 is subject to protein quality control in the nucleus.
Keywords: ERAD; Nuclear membrane; Nuclear proteasome; Protein degradation; Ubiquitylation.
© 2014. Published by The Company of Biologists Ltd.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

