Distinct patterns of diversity, population structure and evolution in the AMA1 genes of sympatric Plasmodium falciparum and Plasmodium vivax populations of Papua New Guinea from an area of similarly high transmission
- PMID: 24930015
- PMCID: PMC4085730
- DOI: 10.1186/1475-2875-13-233
Distinct patterns of diversity, population structure and evolution in the AMA1 genes of sympatric Plasmodium falciparum and Plasmodium vivax populations of Papua New Guinea from an area of similarly high transmission
Abstract
Background: As Plasmodium falciparum and Plasmodium vivax co-exist in most malaria-endemic regions outside sub-Saharan Africa, malaria control strategies in these areas must target both species in order to succeed. Population genetic analyses can predict the effectiveness of interventions including vaccines, by providing insight into patterns of diversity and evolution. The aim of this study was to investigate the population genetics of leading malaria vaccine candidate AMA1 in sympatric P. falciparum and P. vivax populations of Papua New Guinea (PNG), an area of similarly high prevalence (Pf = 22.3 to 38.8%, Pv = 15.3 to 31.8%).
Methods: A total of 72 Pfama1 and 102 Pvama1 sequences were collected from two distinct areas, Madang and Wosera, on the highly endemic PNG north coast.
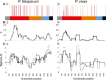
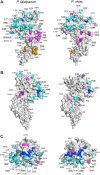
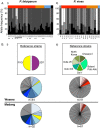
Results: Despite a greater number of polymorphic sites in the AMA1 genes of P. falciparum (Madang = 52; Wosera = 56) compared to P. vivax (Madang = 36, Wosera = 34), the number of AMA1 haplotypes, haplotype diversity (Hd) and recombination (R) was far lower for P. falciparum (Madang = 12, Wosera = 20; Hd ≤0.92, R ≤45.8) than for P. vivax (Madang = 50, Wosera = 38; Hd = 0.99, R = ≤70.9). Balancing selection was detected only within domain I of AMA1 for P. vivax, and in both domains I and III for P. falciparum.
Conclusions: Higher diversity in the genes encoding P. vivax AMA1 than in P. falciparum AMA1 in this highly endemic area has important implications for development of AMA1-based vaccines in PNG and beyond. These results also suggest a smaller effective population size of P. falciparum compared to P. vivax, a finding that warrants further investigation. Differing patterns of selection on the AMA1 genes indicate that critical antigenic sites may differ between the species, highlighting the need for independent investigations of these two leading vaccine candidates.
Figures

References
-
- Barcus MJ, Basri H, Picarima H, Manyakori C, Sekartuti, Elyazar I, Bangs MJ, Maguire JD, Baird JK. Demographic risk factors for severe and fatal vivax and falciparum malaria among hospital admissions in northeastern Indonesian Papua. Am J Trop Med Hyg. 2007;77:984–991. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

