The calcineurin signaling network evolves via conserved kinase-phosphatase modules that transcend substrate identity
- PMID: 24930733
- PMCID: PMC4127121
- DOI: 10.1016/j.molcel.2014.05.012
The calcineurin signaling network evolves via conserved kinase-phosphatase modules that transcend substrate identity
Abstract
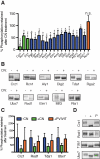
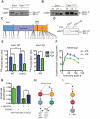
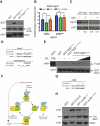
To define a functional network for calcineurin, the conserved Ca(2+)/calmodulin-regulated phosphatase, we systematically identified its substrates in S. cerevisiae using phosphoproteomics and bioinformatics, followed by copurification and dephosphorylation assays. This study establishes new calcineurin functions and reveals mechanisms that shape calcineurin network evolution. Analyses of closely related yeasts show that many proteins were recently recruited to the network by acquiring a calcineurin-recognition motif. Calcineurin substrates in yeast and mammals are distinct due to network rewiring but, surprisingly, are phosphorylated by similar kinases. We postulate that corecognition of conserved substrate features, including phosphorylation and docking motifs, preserves calcineurin-kinase opposition during evolution. One example we document is a composite docking site that confers substrate recognition by both calcineurin and MAPK. We propose that conserved kinase-phosphatase pairs define the architecture of signaling networks and allow other connections between kinases and phosphatases to develop that establish common regulatory motifs in signaling networks.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures




References
-
- Aramburu J, Garcia-Cozar F, Raghavan A, Okamura H, Rao A, Hogan PG. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol Cell. 1998;1:627–637. - PubMed
-
- Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. - PubMed
-
- Asano S, Park JE, Yu LR, Zhou M, Sakchaisri K, Park CJ, Kang YH, Thorner J, Veenstra TD, Lee KS. Direct phosphorylation and activation of a Nim1-related kinase Gin4 by Elm1 in budding yeast. J Biol Chem. 2006;281:27090–27098. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
