Inferring gene ontologies from pairwise similarity data
- PMID: 24932003
- PMCID: PMC4058954
- DOI: 10.1093/bioinformatics/btu282
Inferring gene ontologies from pairwise similarity data
Abstract
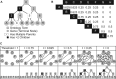
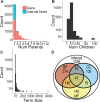
Motivation: While the manually curated Gene Ontology (GO) is widely used, inferring a GO directly from -omics data is a compelling new problem. Recognizing that ontologies are a directed acyclic graph (DAG) of terms and hierarchical relations, algorithms are needed that: analyze a full matrix of gene-gene pairwise similarities from -omics data; infer true hierarchical structure in these data rather than enforcing hierarchy as a computational artifact; and respect biological pleiotropy, by which a term in the hierarchy can relate to multiple higher level terms. Methods addressing these requirements are just beginning to emerge-none has been evaluated for GO inference.
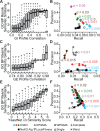
Methods: We consider two algorithms [Clique Extracted Ontology (CliXO), LocalFitness] that uniquely satisfy these requirements, compared with methods including standard clustering. CliXO is a new approach that finds maximal cliques in a network induced by progressive thresholding of a similarity matrix. We evaluate each method's ability to reconstruct the GO biological process ontology from a similarity matrix based on (a) semantic similarities for GO itself or (b) three -omics datasets for yeast.
Results: For task (a) using semantic similarity, CliXO accurately reconstructs GO (>99% precision, recall) and outperforms other approaches (<20% precision, <20% recall). For task (b) using -omics data, CliXO outperforms other methods using two -omics datasets and achieves ∼30% precision and recall using YeastNet v3, similar to an earlier approach (Network Extracted Ontology) and better than LocalFitness or standard clustering (20-25% precision, recall).
Conclusion: This study provides algorithmic foundation for building gene ontologies by capturing hierarchical and pleiotropic structure embedded in biomolecular data.
© The Author 2014. Published by Oxford University Press.
Figures



References
-
- Ahn YY, et al. Link communities reveal multiscale complexity in networks. Nature. 2010;466:761–764. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

