Using association rule mining to determine promising secondary phenotyping hypotheses
- PMID: 24932005
- PMCID: PMC4059059
- DOI: 10.1093/bioinformatics/btu260
Using association rule mining to determine promising secondary phenotyping hypotheses
Abstract
Motivation: Large-scale phenotyping projects such as the Sanger Mouse Genetics project are ongoing efforts to help identify the influences of genes and their modification on phenotypes. Gene-phenotype relations are crucial to the improvement of our understanding of human heritable diseases as well as the development of drugs. However, given that there are ∼: 20 000 genes in higher vertebrate genomes and the experimental verification of gene-phenotype relations requires a lot of resources, methods are needed that determine good candidates for testing.
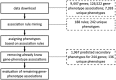
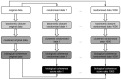
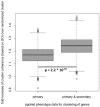
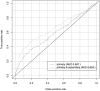
Results: In this study, we applied an association rule mining approach to the identification of promising secondary phenotype candidates. The predictions rely on a large gene-phenotype annotation set that is used to find occurrence patterns of phenotypes. Applying an association rule mining approach, we could identify 1967 secondary phenotype hypotheses that cover 244 genes and 136 phenotypes. Using two automated and one manual evaluation strategies, we demonstrate that the secondary phenotype candidates possess biological relevance to the genes they are predicted for. From the results we conclude that the predicted secondary phenotypes constitute good candidates to be experimentally tested and confirmed.
Availability: The secondary phenotype candidates can be browsed through at http://www.sanger.ac.uk/resources/databases/phenodigm/gene/secondaryphenotype/list.
Contact: ao5@sanger.ac.uk or ds5@sanger.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author 2014. Published by Oxford University Press.
Figures
Similar articles
-
PhenoDigm: analyzing curated annotations to associate animal models with human diseases.Database (Oxford). 2013 May 9;2013:bat025. doi: 10.1093/database/bat025. Print 2013. Database (Oxford). 2013. PMID: 23660285 Free PMC article.
-
Estimating the occurrence of primary ubiquinone deficiency by analysis of large-scale sequencing data.Sci Rep. 2017 Dec 18;7(1):17744. doi: 10.1038/s41598-017-17564-y. Sci Rep. 2017. PMID: 29255295 Free PMC article.
-
Linking tissues to phenotypes using gene expression profiles.Database (Oxford). 2014 Mar 13;2014:bau017. doi: 10.1093/database/bau017. Print 2014. Database (Oxford). 2014. PMID: 24634472 Free PMC article.
-
Clinical syndromes associated with Coenzyme Q10 deficiency.Essays Biochem. 2018 Jul 20;62(3):377-398. doi: 10.1042/EBC20170107. Print 2018 Jul 20. Essays Biochem. 2018. PMID: 30030365 Review.
-
Genetic bases and clinical manifestations of coenzyme Q10 (CoQ 10) deficiency.J Inherit Metab Dis. 2015 Jan;38(1):145-56. doi: 10.1007/s10545-014-9749-9. Epub 2014 Aug 5. J Inherit Metab Dis. 2015. PMID: 25091424 Review.
Cited by
-
PhenoMiner: from text to a database of phenotypes associated with OMIM diseases.Database (Oxford). 2015 Oct 27;2015:bav104. doi: 10.1093/database/bav104. Print 2015. Database (Oxford). 2015. PMID: 26507285 Free PMC article.
-
mtDNA Single-Nucleotide Variants Associated with Type 2 Diabetes.Curr Issues Mol Biol. 2023 Oct 30;45(11):8716-8732. doi: 10.3390/cimb45110548. Curr Issues Mol Biol. 2023. PMID: 37998725 Free PMC article.
-
Usefulness of Vaccine Adverse Event Reporting System for Machine-Learning Based Vaccine Research: A Case Study for COVID-19 Vaccines.Int J Mol Sci. 2022 Jul 26;23(15):8235. doi: 10.3390/ijms23158235. Int J Mol Sci. 2022. PMID: 35897804 Free PMC article.
References
-
- Agrawal R, et al. Fast discovery of association rules. In: Fayyad U, et al., editors. Advances in Knowledge Discovery and Data Mining. Menlo Park, California: AAAI Press; 1996. pp. 307–328.
-
- Amberger J, et al. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM®) Hum. Mutat. 2011;32:564–567. - PubMed
-
- Aymé S. Orphanet, an information site on rare diseases. Soins. 2003;672:46–47. - PubMed
-
- Borgelt C. Workshop of Frequent Item Set Mining Implementations (FIMI 2003) 2003. Efficient implementations of apriori and eclat.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

